Visualize PMP Modes of Variability results: CMIP6
In this document, you will visualize PMP’s Modes of Variability results for CMIP6, to inter-compare performace. You will create following plots.
Portrait plot
Parallel coordinate plot
Written by Jiwoo Lee (LLNL/PCMDI)
Last update: Nov 2024
Contents
Read data from JSON files
1.1 Download PMP output JSON files for CMIP models
1.2 Normalize each column by its median for portrait plot
Portrait Plot
Parallel Coordinate Plot
1. Read data from JSON files
Input data for portrait plot is expected as a set a (stacked or list of) 2-d numpy array(s) with list of strings for x and y axes labels.
1.1 Download PMP output JSON files for CMIP models
[1]:
import glob
import json
import os
import pandas as pd
import numpy as np
from pcmdi_metrics.graphics import download_archived_results
from pcmdi_metrics.utils import sort_human
PMP output files downloadable from the PMP results archive.
[2]:
modes = ['NAM', 'NAO', 'NPGO', 'NPO', 'PDO', 'PNA', 'SAM']
json_dir = './json_files'
mip = "cmip6"
exp = "historical"
data_version = "v20220825"
"""
mip = "cmip5"
exp = "historical"
data_version = "v20210119"
"""
"""
mip = "cmip3"
exp = "20cm3"
data_version = "v20210119"
"""
stat = "rms"
#stat = "rmsc"
#stat = "stdv_pc_ratio_to_obs"
[3]:
if stat == "rms":
stat_name = "RMSE"
elif stat == "rmsc":
stat_name = "Centered RMSE"
elif stat == "stdv_pc_ratio_to_obs":
stat_name = "Amplitude ratio to OBS"
Provide directory path and filename in the PMP results archive.
[4]:
for mode in modes:
if mode in ['PDO', 'NPGO']:
ref = "HadISSTv1.1"
else:
ref = "NOAA-CIRES_20CR"
if mode in ['NPO', 'NPGO']:
eof = "EOF2"
else:
eof = "EOF1"
path = os.path.join("metrics_results/variability_modes/"+mip+"/"+exp+"/"+data_version,
mode, ref,
"_".join(["var", "mode", mode, eof, "stat", mip, exp, "mo_atm_allModels_allRuns_1900-2005.json"]))
download_archived_results(path, json_dir)
Check JSON files
[5]:
json_list = sorted(glob.glob(os.path.join(json_dir, 'var_mode_*' + mip + '*' + '.json')))
for json_file in json_list:
print(json_file.split('/')[-1])
var_mode_NAM_EOF1_stat_cmip6_historical_mo_atm_allModels_allRuns_1900-2005.json
var_mode_NAO_EOF1_stat_cmip6_historical_mo_atm_allModels_allRuns_1900-2005.json
var_mode_NPGO_EOF2_stat_cmip6_historical_mo_atm_allModels_allRuns_1900-2005.json
var_mode_NPO_EOF2_stat_cmip6_historical_mo_atm_allModels_allRuns_1900-2005.json
var_mode_PDO_EOF1_stat_cmip6_historical_mo_atm_allModels_allRuns_1900-2005.json
var_mode_PNA_EOF1_stat_cmip6_historical_mo_atm_allModels_allRuns_1900-2005.json
var_mode_SAM_EOF1_stat_cmip6_historical_mo_atm_allModels_allRuns_1900-2005.json
[6]:
cmip_result_dict = dict()
modes = list()
for json_file in json_list:
mode = json_file.split('/')[-1].split('_')[2]
modes.append(mode)
with open(json_file) as fj:
dict_temp = json.load(fj)['RESULTS']
cmip_result_dict[mode] = dict_temp
[ ]:
def dict_to_df(cmip_result_dict):
models = sorted(list(cmip_result_dict['NAM'].keys()))
df = pd.DataFrame()
df['model'] = models
df['num_runs'] = np.nan
mode_season_list = list()
modes = ['SAM', 'NAM', 'NAO', 'NPO', 'PNA', 'NPGO', 'PDO']
for mode in modes:
if mode in ['PDO', 'NPGO']:
seasons = ['monthly']
else:
seasons = ['DJF', 'MAM', 'JJA', 'SON']
for season in seasons:
df[mode+"_"+season] = np.nan
mode_season_list.append(mode+"_"+season)
for index, model in enumerate(models):
if model in list(cmip_result_dict[mode].keys()):
runs = sort_human(list(cmip_result_dict[mode][model].keys()))
stat_run_list = list()
for run in runs:
stat_run = cmip_result_dict[mode][model][run]['defaultReference'][mode][season]['cbf'][stat]
stat_run_list.append(stat_run)
stat_model = np.average(np.array(stat_run_list))
num_runs = len(runs)
df.at[index, mode+"_"+season] = stat_model
if np.isnan(df.at[index, 'num_runs']):
df.at[index, 'num_runs'] = num_runs
else:
stat_model = np.nan
num_runs = 0
return df, mode_season_list
[8]:
df, mode_season_list = dict_to_df(cmip_result_dict)
df
[8]:
| model | num_runs | SAM_DJF | SAM_MAM | SAM_JJA | SAM_SON | NAM_DJF | NAM_MAM | NAM_JJA | NAM_SON | ... | NPO_DJF | NPO_MAM | NPO_JJA | NPO_SON | PNA_DJF | PNA_MAM | PNA_JJA | PNA_SON | NPGO_monthly | PDO_monthly | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ACCESS-CM2 | 5.0 | 0.574046 | 0.380609 | 0.544973 | 0.391616 | 0.571041 | 0.459388 | 0.240580 | 0.342555 | ... | 0.732129 | 0.460701 | 0.288707 | 0.306164 | 0.650744 | 0.569345 | 0.286248 | 0.256308 | 0.136352 | 0.122066 |
| 1 | ACCESS-ESM1-5 | 40.0 | 0.602834 | 0.414287 | 0.608346 | 0.610213 | 0.506217 | 0.412721 | 0.264908 | 0.354789 | ... | 0.579083 | 0.350249 | 0.269973 | 0.241847 | 0.476215 | 0.388328 | 0.269333 | 0.287768 | 0.126786 | 0.124546 |
| 2 | AWI-CM-1-1-MR | 5.0 | 0.508376 | 0.366660 | 0.796009 | 0.917559 | 0.587488 | 0.392637 | 0.228123 | 0.323303 | ... | 0.682399 | 0.413841 | 0.224981 | 0.406585 | 0.714271 | 0.464686 | 0.276964 | 0.328163 | 0.112087 | 0.136091 |
| 3 | AWI-ESM-1-1-LR | 1.0 | 0.544485 | 0.398040 | 0.634991 | 1.008028 | 0.802982 | 0.475390 | 0.257938 | 0.404807 | ... | 1.065016 | 0.574711 | 0.268740 | 0.307862 | 1.031246 | 0.445994 | 0.295466 | 0.302287 | 0.113582 | 0.194669 |
| 4 | BCC-CSM2-MR | 3.0 | 0.465351 | 0.420144 | 0.624178 | 0.645676 | 0.718848 | 0.728025 | 0.403785 | 0.701120 | ... | 0.763210 | 0.769850 | 0.500015 | 0.591631 | 0.790271 | 0.886669 | 0.483820 | 0.563353 | NaN | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 60 | NorESM2-MM | 3.0 | 0.486581 | 0.399562 | 0.656545 | 0.633588 | 0.548388 | 0.588263 | 0.309797 | 0.411143 | ... | 0.307830 | 0.447543 | 0.384719 | 0.251864 | 0.633810 | 0.720310 | 0.375183 | 0.338101 | 0.105681 | 0.186747 |
| 61 | SAM0-UNICON | 1.0 | 0.511490 | 0.412573 | 0.708228 | 0.679179 | 0.619961 | 0.641307 | 0.233174 | 0.375586 | ... | 0.582020 | 0.343711 | 0.288251 | 0.473160 | 0.684519 | 0.619769 | 0.267318 | 0.387104 | 0.137859 | 0.133782 |
| 62 | TaiESM1 | 2.0 | 0.469439 | 0.392923 | 0.598709 | 0.523395 | 0.669217 | 0.616860 | 0.276326 | 0.450474 | ... | 0.339072 | 0.337362 | 0.336802 | 0.414093 | 0.580974 | 0.631163 | 0.306005 | 0.524750 | 0.137300 | 0.136674 |
| 63 | UKESM1-0-LL | 19.0 | 0.586528 | 0.352492 | 0.513565 | 0.476506 | 0.520315 | 0.506951 | 0.227039 | 0.312856 | ... | 0.673702 | 0.556370 | 0.251275 | 0.250827 | 0.597751 | 0.644911 | 0.240213 | 0.237588 | 0.146209 | 0.145564 |
| 64 | UKESM1-1-LL | 1.0 | 0.565858 | 0.309672 | 0.426744 | 0.498278 | 0.541460 | 0.355031 | 0.265088 | 0.262266 | ... | 0.482643 | 0.409192 | 0.281857 | 0.265768 | 0.480849 | 0.355581 | 0.251933 | 0.164412 | NaN | NaN |
65 rows × 24 columns
[9]:
df_combined = df
[10]:
model_labels = [m + ' (' + str(int(r)) + ')' for m, r in zip(df_combined["model"].to_list(), df_combined["num_runs"].to_list())]
Prepare input for portrait plot plotting function
[11]:
landscape = True
#landscape = False
data = dict()
if landscape:
data = df_combined[mode_season_list].to_numpy().T
else:
data = df_combined[mode_season_list].to_numpy()
[12]:
models = df_combined.index.values.tolist()
print('data.shape:', data.shape)
print('len(mode_season_list): ', len(mode_season_list))
print('len(models): ', len(models))
data.shape: (22, 65)
len(mode_season_list): 22
len(models): 65
[13]:
if landscape:
yaxis_labels = mode_season_list
xaxis_labels = model_labels
else:
xaxis_labels = mode_season_list
yaxis_labels = model_labels
1.2 Normalize each column by its median for portrait plot
Use normalize_by_median function.
Parameters
data: 2d numpy arrayaxis: 0 (normalize each column) or 1 (normalize each row), default=0
Return
data_nor: 2d numpy array
[14]:
if landscape:
axis = 1
figsize = (40, 10)
else:
axis = 0
figsize = (18, 25)
[15]:
from pcmdi_metrics.graphics import normalize_by_median
if stat not in ["stdv_pc_ratio_to_obs"]:
data_nor = normalize_by_median(data, axis=axis)
cmap_bounds = [-0.5, -0.4, -0.3, -0.2, -0.1, 0, 0.1, 0.2, 0.3, 0.4, 0.5]
vertical_center = "median"
cmap = 'RdYlBu_r'
else:
data_nor = data
cmap_bounds = [0.5, 0.7, 0.9, 1.1, 1.3, 1.5]
cmap_bounds = [r/10 for r in range(5, 16, 1)]
vertical_center = 1
cmap = 'jet'
[16]:
data_nor.shape
[16]:
(22, 65)
2. Portrait Plot
Use Matplotlib-based PMP Visualization Function. Detailed description for the functions parameters and returns can be found in the API documentation.
[17]:
from pcmdi_metrics.graphics import portrait_plot
Portrait Plot
[18]:
fig, ax, cbar = portrait_plot(data_nor,
xaxis_labels=xaxis_labels,
yaxis_labels=yaxis_labels,
cbar_label=stat_name,
box_as_square=True,
vrange=(-0.5, 0.5),
figsize=figsize,
cmap=cmap,
cmap_bounds=cmap_bounds,
cbar_kw={"extend": "both"},
missing_color='white',
legend_box_xy=(1.11, 1.21),
legend_box_size=4,
legend_lw=1,
legend_fontsize=15,
logo_rect = [0.67, 1, 0.15, 0.15],
)
ax.set_xticklabels(xaxis_labels, rotation=90, va='center', ha="left")
# Add title
ax.set_title("Variability Modes: "+stat_name, fontsize=30, pad=30)
[18]:
Text(0.5, 1.0, 'Variability Modes: RMSE')
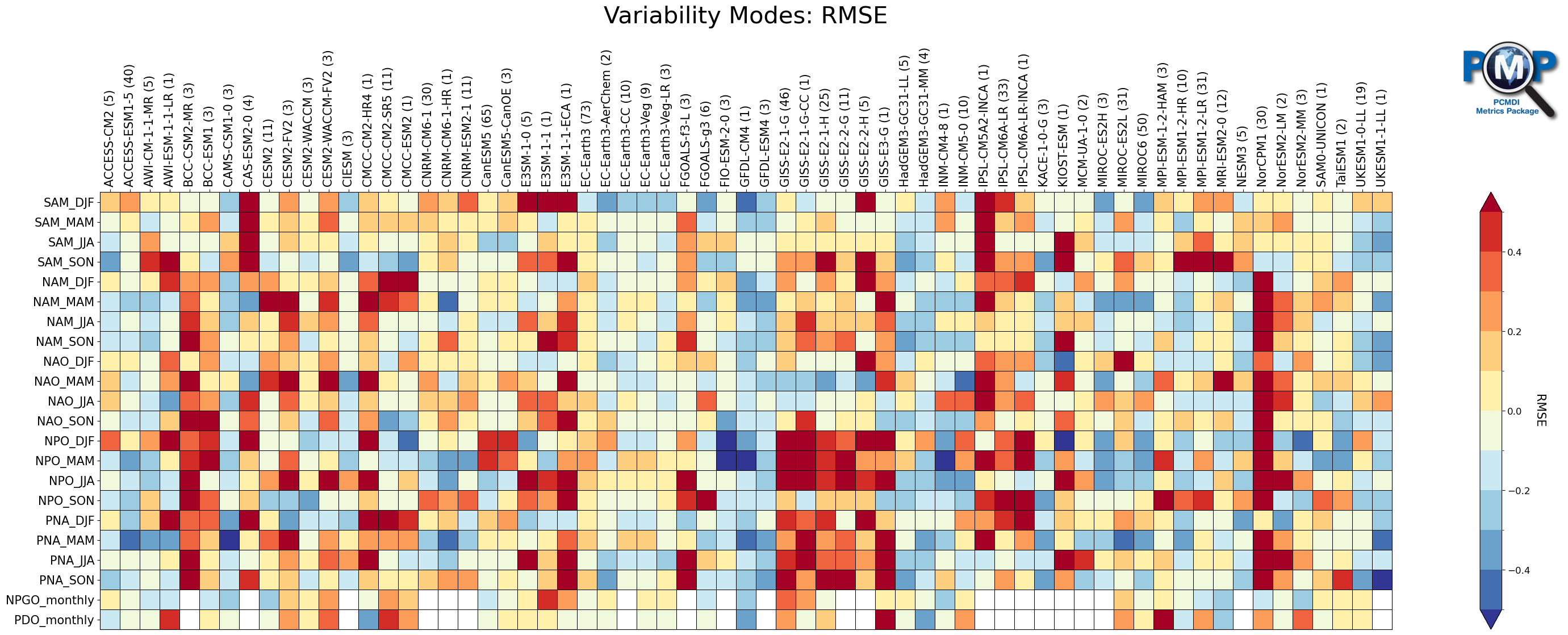
[19]:
# Save figure as an image file
fig.savefig('_'.join(['variability_modes_portrait_plot', mip, exp, stat])+'.png', facecolor='w', bbox_inches='tight')
3. Parallel Coordinate Plot
[20]:
data = df_combined[mode_season_list].to_numpy()
model_names = model_labels
metric_names = mode_season_list
model_highlights = None
print('data.shape:', data.shape)
print('len(metric_names): ', len(metric_names))
print('len(model_names): ', len(model_names))
data.shape: (65, 22)
len(metric_names): 22
len(model_names): 65
Use parallel coordinate plot function of PMP
Detailed description for the functions parameters and returns can be found in the API documentation.
[21]:
from pcmdi_metrics.graphics import parallel_coordinate_plot
3.1 Show all models
[22]:
fig, ax = parallel_coordinate_plot(data, metric_names, model_names,
title='Variability Modes: '+stat_name,
figsize=(21, 7),
colormap='tab20',
show_boxplot=False,
show_violin=True,
violin_colors=("lightgrey", "pink"),
xtick_labelsize=10,
logo_rect=[0.8, 0.8, 0.15, 0.15],
comparing_models=model_highlights,
vertical_center=vertical_center,
vertical_center_line=True
)
ax.set_xticklabels(metric_names, rotation=30, va='top', ha="right")
[22]:
[Text(0, 0, 'SAM_DJF'),
Text(1, 0, 'SAM_MAM'),
Text(2, 0, 'SAM_JJA'),
Text(3, 0, 'SAM_SON'),
Text(4, 0, 'NAM_DJF'),
Text(5, 0, 'NAM_MAM'),
Text(6, 0, 'NAM_JJA'),
Text(7, 0, 'NAM_SON'),
Text(8, 0, 'NAO_DJF'),
Text(9, 0, 'NAO_MAM'),
Text(10, 0, 'NAO_JJA'),
Text(11, 0, 'NAO_SON'),
Text(12, 0, 'NPO_DJF'),
Text(13, 0, 'NPO_MAM'),
Text(14, 0, 'NPO_JJA'),
Text(15, 0, 'NPO_SON'),
Text(16, 0, 'PNA_DJF'),
Text(17, 0, 'PNA_MAM'),
Text(18, 0, 'PNA_JJA'),
Text(19, 0, 'PNA_SON'),
Text(20, 0, 'NPGO_monthly'),
Text(21, 0, 'PDO_monthly')]
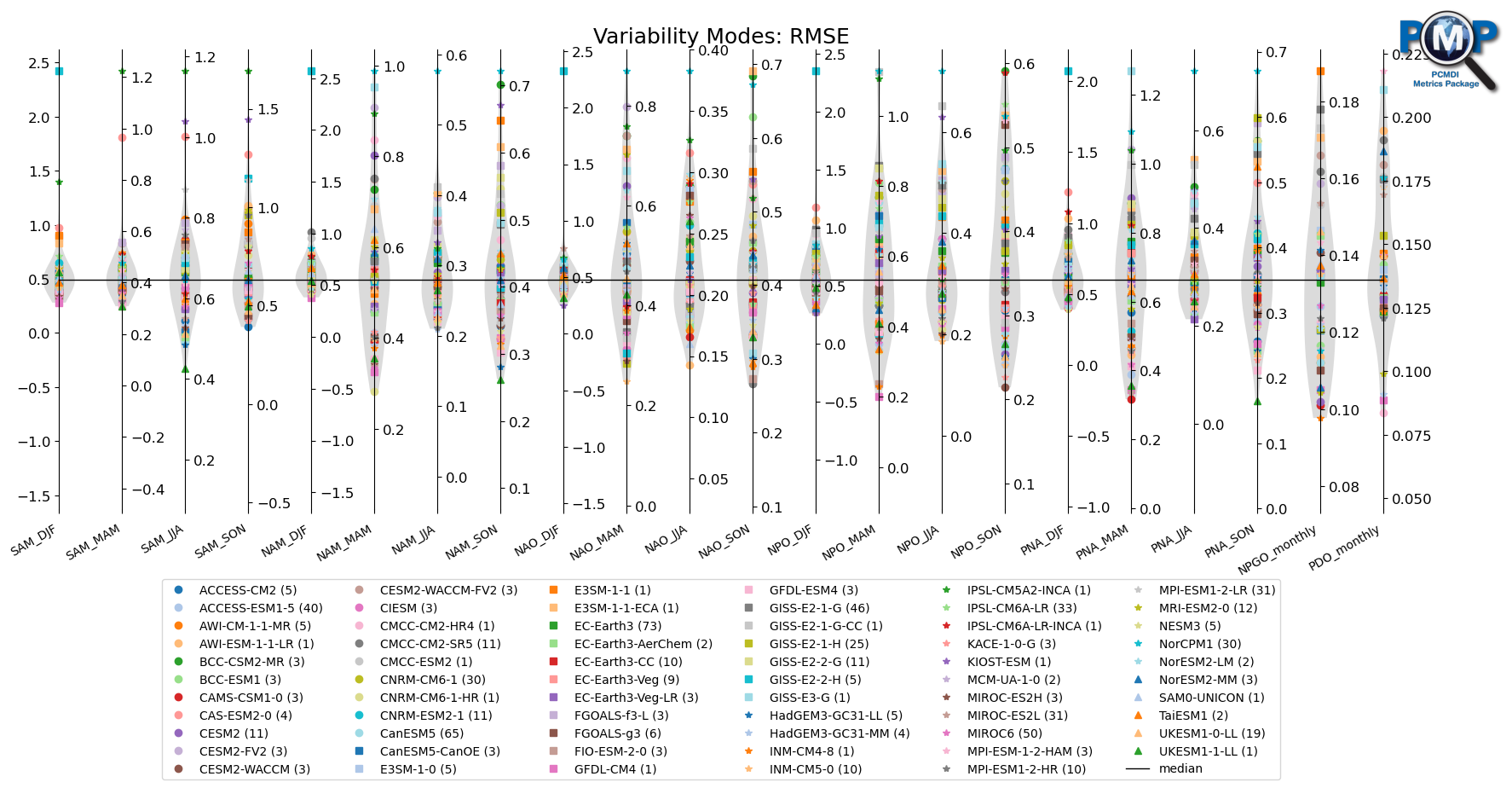
[23]:
# Save figure as an image file
fig.savefig('_'.join(['variability_modes_parallel_coordinate_plot', mip, exp, stat])+'.png', facecolor='w', bbox_inches='tight')
3.2 Highlight specific models
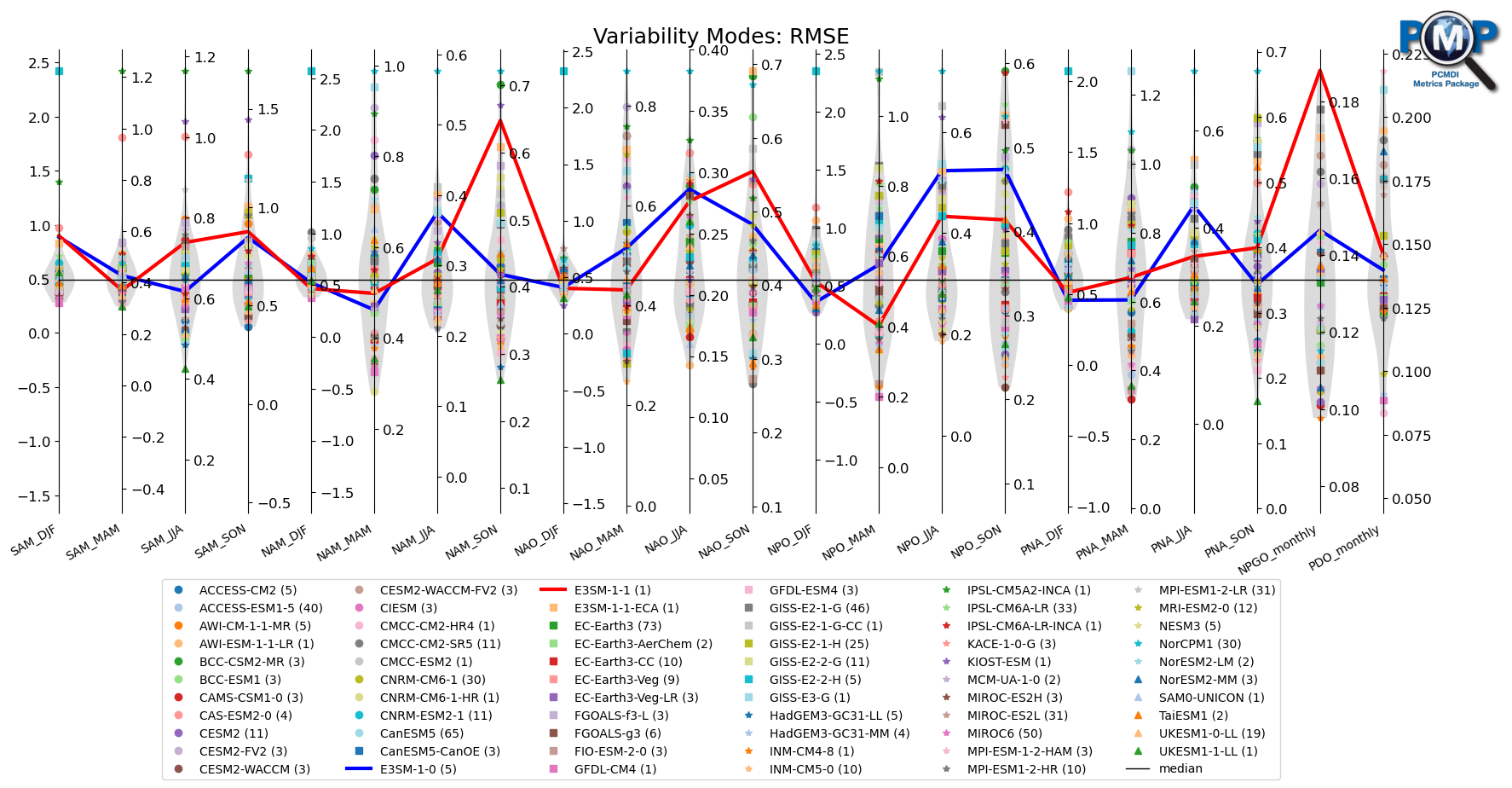
[24]:
model_highlights = ['E3SM-1-0 (5)', 'E3SM-1-1 (1)']
fig, ax = parallel_coordinate_plot(data, metric_names, model_names,
models_to_highlight=model_highlights,
models_to_highlight_colors=['blue', 'red'],
title='Variability Modes: '+stat_name,
figsize=(21, 7),
colormap='tab20',
show_boxplot=False,
show_violin=True,
violin_colors=("lightgrey", "pink"),
xtick_labelsize=10,
logo_rect=[0.8, 0.8, 0.15, 0.15],
comparing_models=model_highlights,
vertical_center=vertical_center,
vertical_center_line=True
)
ax.set_xticklabels(metric_names, rotation=30, va='top', ha="right")
[24]:
[Text(0, 0, 'SAM_DJF'),
Text(1, 0, 'SAM_MAM'),
Text(2, 0, 'SAM_JJA'),
Text(3, 0, 'SAM_SON'),
Text(4, 0, 'NAM_DJF'),
Text(5, 0, 'NAM_MAM'),
Text(6, 0, 'NAM_JJA'),
Text(7, 0, 'NAM_SON'),
Text(8, 0, 'NAO_DJF'),
Text(9, 0, 'NAO_MAM'),
Text(10, 0, 'NAO_JJA'),
Text(11, 0, 'NAO_SON'),
Text(12, 0, 'NPO_DJF'),
Text(13, 0, 'NPO_MAM'),
Text(14, 0, 'NPO_JJA'),
Text(15, 0, 'NPO_SON'),
Text(16, 0, 'PNA_DJF'),
Text(17, 0, 'PNA_MAM'),
Text(18, 0, 'PNA_JJA'),
Text(19, 0, 'PNA_SON'),
Text(20, 0, 'NPGO_monthly'),
Text(21, 0, 'PDO_monthly')]