6. ENSO
This notebook provides an overview of running the ENSO Metrics package through the PMP.
Reference
Planton, Y., E. Guilyardi, A. T. Wittenberg, J. Lee, P. J. Gleckler, T. Bayr, S. McGregor, M. J. McPhaden, S. Power, R. Roehrig, A. Voldoire, 2020: Evaluating El Niño in climate models with the CLIVAR 2020 ENSO metrics package. Bulletin of the American Meteorological Society. doi: 10.1175/BAMS-D-19-0337.1
Lee, J., P. J. Gleckler, M.-S. Ahn, A. Ordonez, P. Ullrich, K. R. Sperber, K. E. Taylor, Y. Y. Planton, E. Guilyardi, P. Durack, C. Bonfils, M. D. Zelinka, L.-W. Chao, B. Dong, C. Doutriaux, C. Zhang, T. Vo, J. Boutte, M. F. Wehner, A. G. Pendergrass, D. Kim, Z. Xue, A. T. Wittenberg, and J. Krasting, 2024: Systematic and Objective Evaluation of Earth System Models: PCMDI Metrics Package (PMP) version 3. Geoscientific Model Development, 17, 3919–3948, doi: 10.5194/gmd-17-3919-2024
Description for individual metrics can be found at https://github.com/CLIVAR-PRP/ENSO_metrics/wiki.
If enso_package is not installed in your environment, you will need to install it.
To check whether the ENSO metrics package is installed in the current virtual conda environment:
conda list enso_metrics
To install the ENSO metrics package in the current virtual conda environment:
conda install -c conda-forge enso_metrics
Unquote the commend in the following box to install the ENSO package. Further installation instruction is available here: https://github.com/CLIVAR-PRP/ENSO_metrics/wiki/install
Please note, ENSO package requires Python 3.10.x.
[ ]:
"""
!conda install -c conda-forge enso_metrics
"""
Download demo data
The ENSO metrics demo requires downloading a large sample data set (size 10.8 GB). The ENSO metric requires a different set of sample data than the rest of the PMP metrics. This section of the notebook will download that data to your chosen location and generate a basic parameter file.
[1]:
# Lets get the file containing the data needed for this demo
import requests
r = requests.get("https://pcmdiweb.llnl.gov/pss/pmpdata/pmp_enso_tutorial_files.v20210823.txt")
with open("enso_data_files.txt","wb") as f:
f.write(r.content)
If you want to change the location where the demo data and output are stored, you can do so here:
[2]:
# This is where you will be downloading the sample_data
demo_data_directory = "demo_data"
# this line is where your output will be stored
demo_output_directory = "demo_output"
Then download the data. The total sample data size is 10.8 GB. This will take several minutes.
[3]:
# Let's download the files
from pcmdi_metrics.io.base import download_sample_data_files
try:
download_sample_data_files("enso_data_files.txt", demo_data_directory)
print("All files downloaded")
except:
print("Download failed")
All files downloaded
After downloading the data, we generate the parameter file for this demo.
[4]:
from download_sample_data import generate_parameter_files
filenames=["basic_enso_param.py.in"]
generate_parameter_files(demo_data_directory, demo_output_directory, filenames=filenames)
Preparing parameter file: basic_enso_param.py
Saving User Choices
[5]:
# To open and display one of the graphics
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
from matplotlib import rcParams
import os
%matplotlib inline
Usage
The ENSO driver can be run from the command line as enso_driver.py. In this notebook, we will use bash cell magic (cells beginning with %%bash) to run the ENSO driver as a subprocess.
For help, type:
enso_driver.py --help
[6]:
%%bash
enso_driver.py --help
usage: enso_driver.py [-h] [--parameters PARAMETERS]
[--diags OTHER_PARAMETERS [OTHER_PARAMETERS ...]]
[--mip MIP] [--exp EXP] [--modpath MODPATH]
[--modpath_lf MODPATH_LF]
[--modnames MODNAMES [MODNAMES ...]] [-r REALIZATION]
[--reference_data_path REFERENCE_DATA_PATH]
[--reference_data_lf_path REFERENCE_DATA_LF_PATH]
[--metricsCollection METRICSCOLLECTION]
[--json_name JSON_NAME] [--netcdf_name NETCDF_NAME]
[--results_dir RESULTS_DIR] [--case_id CASE_ID]
[--obs_catalogue OBS_CATALOGUE]
[--obs_cmor_path OBS_CMOR_PATH] [-d [DEBUG]]
[--obs_cmor [OBS_CMOR]] [--nc_out [NC_OUT]]
options:
-h, --help show this help message and exit
--parameters PARAMETERS, -p PARAMETERS
--diags OTHER_PARAMETERS [OTHER_PARAMETERS ...]
Path to other user-defined parameter file. (default:
None)
--mip MIP A WCRP MIP project such as CMIP3 and CMIP5 (default:
cmip5)
--exp EXP An experiment such as AMIP, historical or pi-contorl
(default: historical)
--modpath MODPATH, --mp MODPATH
Explicit path to model data (default: None)
--modpath_lf MODPATH_LF
Directory path to model land fraction field (default:
None)
--modnames MODNAMES [MODNAMES ...]
List of models (default: None)
-r REALIZATION, --realization REALIZATION
Consider all accessible realizations as idividual -
r1i1p1: default, consider only 'r1i1p1' member Or,
specify realization, e.g, r3i1p1' - *: consider all
available realizations (default: r1i1p1)
--reference_data_path REFERENCE_DATA_PATH, --rdp REFERENCE_DATA_PATH
The path/filename of reference (obs) data. (default:
None)
--reference_data_lf_path REFERENCE_DATA_LF_PATH
Data path to land fraction of reference dataset
(default: None)
--metricsCollection METRICSCOLLECTION
Metrics Collection e.g. ENSO_perf, ENSO_tel, or
ENSO_proc (default: ENSO_perf)
--json_name JSON_NAME
File name for output JSON (default: None)
--netcdf_name NETCDF_NAME
File name for output NetCDF (default: None)
--results_dir RESULTS_DIR, --rd RESULTS_DIR
The name of the folder where all runs will be stored.
(default: None)
--case_id CASE_ID version as date, e.g., v20191116 (yyyy-mm-dd)
(default: v20250829)
--obs_catalogue OBS_CATALOGUE
obs_catalogue JSON file for CMORized observation,
default is None (default: None)
--obs_cmor_path OBS_CMOR_PATH
Directory path for CMORized observation dataset,
default is None (default: None)
-d [DEBUG], --debug [DEBUG]
Option for debug: True / False (defualt) (default:
False)
--obs_cmor [OBS_CMOR]
Use CMORized reference database?: True / False
(defualt) (default: False)
--nc_out [NC_OUT] Option for generate netCDF file output: True (default)
/ False (default: True)
Basic example
Parameters for the ENSO Metrics can be set on the command line or using a parameter file. This first example will use a parameter file, which is shown below.
[7]:
with open("basic_enso_param.py") as f:
print(f.read())
import os
#
# OPTIONS ARE SET BY USER IN THIS FILE AS INDICATED BELOW BY:
#
#
# MODELS
modnames = ['ACCESS1-0']
mip = 'cmip5' # cmip5, cmip6
exp = 'historical' # historical, piControl
realization = 'r1i1p1'
modpath = 'demo_data/CMIP5_demo_data/%(variable)_Amon_%(model)_historical_%(realization)_185001-200512.nc'
modpath_lf = 'demo_data/CMIP5_demo_data/sftlf_fx_%(model)_amip_r0i0p0.nc'
# OBSERVATIONS
obs_cmor = True
obs_cmor_path = "demo_data/obs4MIPs_PCMDI_monthly"
obs_catalogue = "demo_data/obs4MIPs_PCMDI-CEM2021_monthly_bySource_catalogue_v20210805_demo.json"
# METRICS COLLECTION
metricsCollection = 'ENSO_perf' # ENSO_perf, ENSO_tel, ENSO_proc
# OUTPUT
case_id = 'basicTestEnso'
results_dir = os.path.join('demo_output',case_id, metricsCollection)
json_name = '%(mip)_%(exp)_%(metricsCollection)_%(case_id)_%(model)_%(realization)'
netcdf_name = json_name
nc_out = True
The next cell runs the ENSO driver using the basic parameter file. This may take several minutes.
[8]:
%%bash
enso_driver.py -p basic_enso_param.py
mip: cmip5
exp: historical
models: ['ACCESS1-0']
realization: r1i1p1
mc_name: ENSO_perf
outdir: demo_output/basicTestEnso/ENSO_perf
netcdf_path: demo_output/basicTestEnso/ENSO_perf
debug: False
obs_cmor: True
obs_cmor_path: demo_data/obs4MIPs_PCMDI_monthly
egg_pth: /Users/lee1043/miniforge3/envs/pmp_devel_20250715/share/pmp
output directory for graphics:demo_output/basicTestEnso/ENSO_perf
output directory for diagnostic_results:demo_output/basicTestEnso/ENSO_perf
output directory for metrics_results:demo_output/basicTestEnso/ENSO_perf
list_variables: ['pr', 'sst', 'taux']
list_obs: ['AVISO-1-0', 'ERA-INT', 'GPCP-2-3', 'HadISST-1-1']
PMPdriver: dict_obs readin end
Process start:Fri Aug 29 15:21:52 2025
models: ['ACCESS1-0']
----- model: ACCESS1-0 ---------------------
PMPdriver: var loop start for model ACCESS1-0
realization: r1i1p1
--- run: r1i1p1 ---
--- var: pr ---
var_in_file: pr
var, areacell_in_file, realm: pr areacella atmos
path: demo_data/CMIP5_demo_data/pr_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc
file_list: ['demo_data/CMIP5_demo_data/pr_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
PMPdriver: var loop end
--- var: sst ---
var_in_file: ts
var, areacell_in_file, realm: sst areacella atmos
path: demo_data/CMIP5_demo_data/ts_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc
file_list: ['demo_data/CMIP5_demo_data/ts_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
PMPdriver: var loop end
--- var: taux ---
var_in_file: tauu
var, areacell_in_file, realm: taux areacella atmos
path: demo_data/CMIP5_demo_data/tauu_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc
file_list: ['demo_data/CMIP5_demo_data/tauu_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
PMPdriver: var loop end
dictDatasets:
{
"model": {
"ACCESS1-0_r1i1p1": {
"pr": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/CMIP5_demo_data/pr_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc",
"path + filename_area": "demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"path + filename_landmask": "demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"varname": "pr"
},
"sst": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/CMIP5_demo_data/ts_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc",
"path + filename_area": "demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"path + filename_landmask": "demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"varname": "ts"
},
"taux": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/CMIP5_demo_data/tauu_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc",
"path + filename_area": "demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"path + filename_landmask": "demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"varname": "tauu"
}
}
},
"observations": {
"ERA-Interim": {
"pr": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/obs4MIPs_PCMDI_monthly/ECMWF/ERA-INT/mon/pr/gn/v20210727/pr_mon_ERA-INT_PCMDI_gn_197901-201903.nc",
"path + filename_area": null,
"path + filename_landmask": null,
"varname": "pr"
},
"sst": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/obs4MIPs_PCMDI_monthly/ECMWF/ERA-INT/mon/ts/gn/v20210727/ts_mon_ERA-INT_PCMDI_gn_197901-201903.nc",
"path + filename_area": null,
"path + filename_landmask": null,
"varname": "ts"
},
"taux": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/obs4MIPs_PCMDI_monthly/ECMWF/ERA-INT/mon/tauu/gn/v20210727/tauu_mon_ERA-INT_PCMDI_gn_197901-201903.nc",
"path + filename_area": null,
"path + filename_landmask": null,
"varname": "tauu"
}
},
"GPCPv2.3": {
"pr": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/obs4MIPs_PCMDI_monthly/NOAA-NCEI/GPCP-2-3/mon/pr/gn/v20210727/pr_mon_GPCP-2-3_PCMDI_gn_197901-201907.nc",
"path + filename_area": null,
"path + filename_landmask": null,
"varname": "pr"
}
},
"HadISST": {
"sst": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/obs4MIPs_PCMDI_monthly/MOHC/HadISST-1-1/mon/ts/gn/v20210727/ts_mon_HadISST-1-1_PCMDI_gn_187001-201907.nc",
"path + filename_area": null,
"path + filename_landmask": null,
"varname": "ts"
}
}
}
}
### Compute the metric collection ###
ComputeCollection: metric = BiasPrLatRmse
ComputeMetric: oneVarRMSmetric, BiasPrLatRmse = ACCESS1-0_r1i1p1 and ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = BiasPrLonRmse
ComputeMetric: oneVarRMSmetric, BiasPrLonRmse = ACCESS1-0_r1i1p1 and ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = BiasSstLonRmse
ComputeMetric: oneVarRMSmetric, BiasSstLonRmse = ACCESS1-0_r1i1p1 and ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = BiasTauxLonRmse
ComputeMetric: oneVarRMSmetric, BiasTauxLonRmse = ACCESS1-0_r1i1p1 and ERA-Interim
EnsoUvcdatToolsLib AverageHorizontal
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoAmpl
ComputeMetric: oneVarmetric = ACCESS1-0_r1i1p1
EnsoUvcdatToolsLib AverageMeridional
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/cdms2/dataset.py:2179: Warning: Files are written with compression and no shuffling
You can query different values of compression using the functions:
cdms2.getNetcdfShuffleFlag() returning 1 if shuffling is enabled, 0 otherwise
cdms2.getNetcdfDeflateFlag() returning 1 if deflate is used, 0 otherwise
cdms2.getNetcdfDeflateLevelFlag() returning the level of compression for the deflate method
If you want to turn that off or set different values of compression use the functions:
value = 0
cdms2.setNetcdfShuffleFlag(value) ## where value is either 0 or 1
cdms2.setNetcdfDeflateFlag(value) ## where value is either 0 or 1
cdms2.setNetcdfDeflateLevelFlag(value) ## where value is a integer between 0 and 9 included
To produce NetCDF3 Classic files use:
cdms2.useNetCDF3()
To Force NetCDF4 output with classic format and no compressing use:
cdms2.setNetcdf4Flag(1)
NetCDF4 file with no shuffling or deflate and noclassic will be open for parallel i/o
warnings.warn("Files are written with compression and no shuffling\n" +
ComputeMetric: oneVarmetric = ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoDuration
ComputeMetric: oneVarmetric = ACCESS1-0_r1i1p1
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/cdms2/MV2.py:318: Warning: arguments order for compress function has changed
it is now: MV2.copmress(array,condition), if your code seems to not react or act wrong to a call to compress, please check this
warnings.warn(
ComputeMetric: oneVarmetric = ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoSeasonality
ComputeMetric: oneVarmetric = ACCESS1-0_r1i1p1
EnsoUvcdatToolsLib AverageMeridional
EnsoUvcdatToolsLib AverageMeridional
EnsoUvcdatToolsLib AverageMeridional
ComputeMetric: oneVarmetric = ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoSstDiversity_2
ComputeMetric: oneVarmetric = ACCESS1-0_r1i1p1
EnsoUvcdatToolsLib AverageMeridional
ComputeMetric: oneVarmetric = ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoSstLonRmse
ComputeMetric: oneVarRMSmetric, EnsoSstLonRmse = ACCESS1-0_r1i1p1 and ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoSstSkew
ComputeMetric: oneVarmetric = ACCESS1-0_r1i1p1
EnsoUvcdatToolsLib AverageMeridional
ComputeMetric: oneVarmetric = ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoSstTsRmse
ComputeMetric: oneVarRMSmetric, EnsoSstTsRmse = ACCESS1-0_r1i1p1 and ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = SeasonalPrLatRmse
ComputeMetric: oneVarRMSmetric, SeasonalPrLatRmse = ACCESS1-0_r1i1p1 and ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = SeasonalPrLonRmse
ComputeMetric: oneVarRMSmetric, SeasonalPrLonRmse = ACCESS1-0_r1i1p1 and ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = SeasonalSstLonRmse
ComputeMetric: oneVarRMSmetric, SeasonalSstLonRmse = ACCESS1-0_r1i1p1 and ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = SeasonalTauxLonRmse
ComputeMetric: oneVarRMSmetric, SeasonalTauxLonRmse = ACCESS1-0_r1i1p1 and ERA-Interim
EnsoUvcdatToolsLib AverageHorizontal
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
INFO::2025-08-29 15:23::pcmdi_metrics:: Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_perf/cmip5_historical_ENSO_perf_basicTestEnso_ACCESS1-0_r1i1p1.json
2025-08-29 15:23:13,255 [INFO]: base.py(write:347) >> Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_perf/cmip5_historical_ENSO_perf_basicTestEnso_ACCESS1-0_r1i1p1.json
2025-08-29 15:23:13,255 [INFO]: base.py(write:347) >> Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_perf/cmip5_historical_ENSO_perf_basicTestEnso_ACCESS1-0_r1i1p1.json
INFO::2025-08-29 15:23::pcmdi_metrics:: Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_perf/cmip5_historical_ENSO_perf_basicTestEnso_ACCESS1-0_r1i1p1_diveDown.json
2025-08-29 15:23:18,085 [INFO]: base.py(write:347) >> Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_perf/cmip5_historical_ENSO_perf_basicTestEnso_ACCESS1-0_r1i1p1_diveDown.json
2025-08-29 15:23:18,085 [INFO]: base.py(write:347) >> Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_perf/cmip5_historical_ENSO_perf_basicTestEnso_ACCESS1-0_r1i1p1_diveDown.json
figure plotting start
metrics: ['BiasPrLatRmse', 'BiasPrLonRmse', 'BiasSstLonRmse', 'BiasTauxLonRmse', 'EnsoAmpl', 'EnsoDuration', 'EnsoSeasonality', 'EnsoSstDiversity_2', 'EnsoSstLonRmse', 'EnsoSstSkew', 'EnsoSstTsRmse', 'SeasonalPrLatRmse', 'SeasonalPrLonRmse', 'SeasonalSstLonRmse', 'SeasonalTauxLonRmse']
filename_js: demo_output/basicTestEnso/ENSO_perf/cmip5_historical_ENSO_perf_basicTestEnso_ACCESS1-0_r1i1p1.json
met: BiasPrLatRmse
filename_nc: demo_output/basicTestEnso/ENSO_perf/cmip5_historical_ENSO_perf_basicTestEnso_ACCESS1-0_r1i1p1_BiasPrLatRmse.nc
failed for ACCESS1-0 r1i1p1
'BiasPrLatRmse'
PMPdriver: model loop end
Process end: Fri Aug 29 15:23:18 2025
This run saved metrics to two files:
basicTestEnso/ENSO_perf/cmip5_historical_ENSO_perf_basicTestEnso_ACCESS1-0_r1i1p1.jsonbasicTestEnso/ENSO_perf/cmip5_historical_ENSO_perf_basicTestEnso_ACCESS1-0_r1i1p1_diveDown.json
diveDown metrics are not available in all cases.
Example dive down (i.e., diagnostics) figures:
[9]:
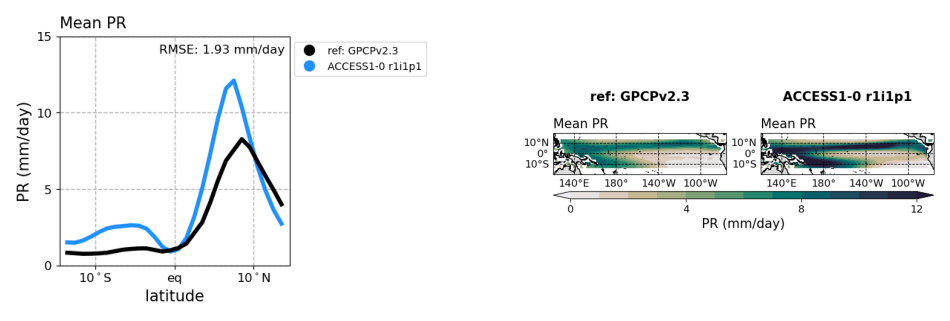
# figure size in inches optional
rcParams['figure.figsize'] = 12, 10
# path to images
plot1 = os.path.join(demo_output_directory,"basicTestEnso/ENSO_perf/cmip5_historical_ENSO_perf_ACCESS1-0_r1i1p1_BiasPrLatRmse_diagnostic_divedown01.png")
plot2 = os.path.join(demo_output_directory,"basicTestEnso/ENSO_perf/cmip5_historical_ENSO_perf_ACCESS1-0_r1i1p1_BiasPrLatRmse_diagnostic_divedown02.png")
# display images
fig, ax = plt.subplots(1,2); ax[0].axis('off'); ax[1].axis('off')
ax[0].imshow(mpimg.imread(plot1))
ax[1].imshow(mpimg.imread(plot2))
[9]:
<matplotlib.image.AxesImage at 0x1affcb610>
The results section of cmip5_historical_ENSO_perf_basicTestEnso_ACCESS1-0_r1i1p1.json is shown below.
[10]:
import json
metrics_file=demo_output_directory+"/basicTestEnso/ENSO_perf/cmip5_historical_ENSO_perf_basicTestEnso_ACCESS1-0_r1i1p1.json"
with open(metrics_file) as f:
results = json.load(f)["RESULTS"]["model"]["ACCESS1-0"]["r1i1p1"]["value"]
print(json.dumps(results, indent = 2))
{}
ENSO Metrics Collections
They can be selected using the --metricsCollection flag. The first example used the “ENSO_perf” collection.
The next example runs the teleconnection collection. To save individual metrics in netCDF format, it uses the --nc_out flag.
[11]:
%%bash -s "$demo_output_directory"
enso_driver.py -p basic_enso_param.py \
--metricsCollection ENSO_tel \
--results_dir $1/basicTestEnso/ENSO_tel \
--nc_out True
mip: cmip5
exp: historical
models: ['ACCESS1-0']
realization: r1i1p1
mc_name: ENSO_tel
outdir: demo_output/basicTestEnso/ENSO_tel
netcdf_path: demo_output/basicTestEnso/ENSO_tel
debug: False
obs_cmor: True
obs_cmor_path: demo_data/obs4MIPs_PCMDI_monthly
egg_pth: /Users/lee1043/miniforge3/envs/pmp_devel_20250715/share/pmp
output directory for graphics:demo_output/basicTestEnso/ENSO_tel
output directory for diagnostic_results:demo_output/basicTestEnso/ENSO_tel
output directory for metrics_results:demo_output/basicTestEnso/ENSO_tel
list_variables: ['pr', 'sst']
list_obs: ['AVISO-1-0', 'ERA-INT', 'GPCP-2-3', 'HadISST-1-1']
PMPdriver: dict_obs readin end
Process start:Fri Aug 29 15:23:25 2025
models: ['ACCESS1-0']
----- model: ACCESS1-0 ---------------------
PMPdriver: var loop start for model ACCESS1-0
realization: r1i1p1
--- run: r1i1p1 ---
--- var: pr ---
var_in_file: pr
var, areacell_in_file, realm: pr areacella atmos
path: demo_data/CMIP5_demo_data/pr_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc
file_list: ['demo_data/CMIP5_demo_data/pr_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
PMPdriver: var loop end
--- var: sst ---
var_in_file: ts
var, areacell_in_file, realm: sst areacella atmos
path: demo_data/CMIP5_demo_data/ts_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc
file_list: ['demo_data/CMIP5_demo_data/ts_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
PMPdriver: var loop end
dictDatasets:
{
"model": {
"ACCESS1-0_r1i1p1": {
"pr": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/CMIP5_demo_data/pr_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc",
"path + filename_area": "demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"path + filename_landmask": "demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"varname": "pr"
},
"sst": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/CMIP5_demo_data/ts_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc",
"path + filename_area": "demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"path + filename_landmask": "demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"varname": "ts"
}
}
},
"observations": {
"ERA-Interim": {
"pr": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/obs4MIPs_PCMDI_monthly/ECMWF/ERA-INT/mon/pr/gn/v20210727/pr_mon_ERA-INT_PCMDI_gn_197901-201903.nc",
"path + filename_area": null,
"path + filename_landmask": null,
"varname": "pr"
},
"sst": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/obs4MIPs_PCMDI_monthly/ECMWF/ERA-INT/mon/ts/gn/v20210727/ts_mon_ERA-INT_PCMDI_gn_197901-201903.nc",
"path + filename_area": null,
"path + filename_landmask": null,
"varname": "ts"
}
},
"GPCPv2.3": {
"pr": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/obs4MIPs_PCMDI_monthly/NOAA-NCEI/GPCP-2-3/mon/pr/gn/v20210727/pr_mon_GPCP-2-3_PCMDI_gn_197901-201907.nc",
"path + filename_area": null,
"path + filename_landmask": null,
"varname": "pr"
}
},
"HadISST": {
"sst": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/obs4MIPs_PCMDI_monthly/MOHC/HadISST-1-1/mon/ts/gn/v20210727/ts_mon_HadISST-1-1_PCMDI_gn_187001-201907.nc",
"path + filename_area": null,
"path + filename_landmask": null,
"varname": "ts"
}
}
}
}
### Compute the metric collection ###
ComputeCollection: metric = EnsoAmpl
ComputeMetric: oneVarmetric = ACCESS1-0_r1i1p1
EnsoUvcdatToolsLib AverageMeridional
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/cdms2/dataset.py:2179: Warning: Files are written with compression and no shuffling
You can query different values of compression using the functions:
cdms2.getNetcdfShuffleFlag() returning 1 if shuffling is enabled, 0 otherwise
cdms2.getNetcdfDeflateFlag() returning 1 if deflate is used, 0 otherwise
cdms2.getNetcdfDeflateLevelFlag() returning the level of compression for the deflate method
If you want to turn that off or set different values of compression use the functions:
value = 0
cdms2.setNetcdfShuffleFlag(value) ## where value is either 0 or 1
cdms2.setNetcdfDeflateFlag(value) ## where value is either 0 or 1
cdms2.setNetcdfDeflateLevelFlag(value) ## where value is a integer between 0 and 9 included
To produce NetCDF3 Classic files use:
cdms2.useNetCDF3()
To Force NetCDF4 output with classic format and no compressing use:
cdms2.setNetcdf4Flag(1)
NetCDF4 file with no shuffling or deflate and noclassic will be open for parallel i/o
warnings.warn("Files are written with compression and no shuffling\n" +
ComputeMetric: oneVarmetric = ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoPrMapDjf
ComputeMetric: twoVarRMSmetric, EnsoPrMapDjf = ACCESS1-0_r1i1p1 and ERA-Interim_ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoPrMapJja
ComputeMetric: twoVarRMSmetric, EnsoPrMapJja = ACCESS1-0_r1i1p1 and ERA-Interim_ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoSeasonality
ComputeMetric: oneVarmetric = ACCESS1-0_r1i1p1
EnsoUvcdatToolsLib AverageMeridional
EnsoUvcdatToolsLib AverageMeridional
EnsoUvcdatToolsLib AverageMeridional
ComputeMetric: oneVarmetric = ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoSstLonRmse
ComputeMetric: oneVarRMSmetric, EnsoSstLonRmse = ACCESS1-0_r1i1p1 and ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoSstMapDjf
ComputeMetric: oneVarRMSmetric, EnsoSstMapDjf = ACCESS1-0_r1i1p1 and ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoSstMapJja
ComputeMetric: oneVarRMSmetric, EnsoSstMapJja = ACCESS1-0_r1i1p1 and ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
INFO::2025-08-29 15:23::pcmdi_metrics:: Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_tel/cmip5_historical_ENSO_tel_basicTestEnso_ACCESS1-0_r1i1p1.json
2025-08-29 15:23:53,700 [INFO]: base.py(write:347) >> Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_tel/cmip5_historical_ENSO_tel_basicTestEnso_ACCESS1-0_r1i1p1.json
2025-08-29 15:23:53,700 [INFO]: base.py(write:347) >> Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_tel/cmip5_historical_ENSO_tel_basicTestEnso_ACCESS1-0_r1i1p1.json
INFO::2025-08-29 15:23::pcmdi_metrics:: Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_tel/cmip5_historical_ENSO_tel_basicTestEnso_ACCESS1-0_r1i1p1_diveDown.json
2025-08-29 15:23:59,279 [INFO]: base.py(write:347) >> Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_tel/cmip5_historical_ENSO_tel_basicTestEnso_ACCESS1-0_r1i1p1_diveDown.json
2025-08-29 15:23:59,279 [INFO]: base.py(write:347) >> Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_tel/cmip5_historical_ENSO_tel_basicTestEnso_ACCESS1-0_r1i1p1_diveDown.json
figure plotting start
metrics: ['EnsoAmpl', 'EnsoPrMapDjf', 'EnsoPrMapJja', 'EnsoSeasonality', 'EnsoSstLonRmse', 'EnsoSstMapDjf', 'EnsoSstMapJja']
filename_js: demo_output/basicTestEnso/ENSO_tel/cmip5_historical_ENSO_tel_basicTestEnso_ACCESS1-0_r1i1p1.json
met: EnsoAmpl
filename_nc: demo_output/basicTestEnso/ENSO_tel/cmip5_historical_ENSO_tel_basicTestEnso_ACCESS1-0_r1i1p1_EnsoAmpl.nc
failed for ACCESS1-0 r1i1p1
'EnsoAmpl'
PMPdriver: model loop end
Process end: Fri Aug 29 15:23:59 2025
All of the results (netCDF and JSON) are located in the output directory, which uses the metrics collection name.
[12]:
!ls {demo_output_directory + "/basicTestEnso/ENSO_tel/*.nc"}
demo_output/basicTestEnso/ENSO_tel/cmip5_historical_ENSO_tel_basicTestEnso_ACCESS1-0_r1i1p1_EnsoAmpl.nc
demo_output/basicTestEnso/ENSO_tel/cmip5_historical_ENSO_tel_basicTestEnso_ACCESS1-0_r1i1p1_EnsoSeasonality.nc
[13]:
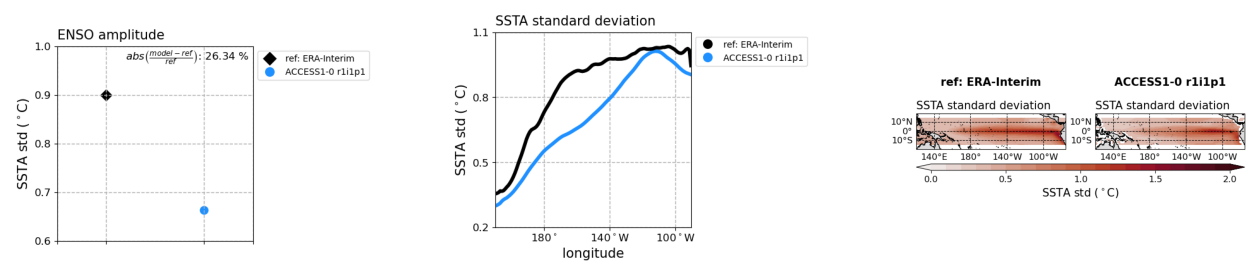
# figure size in inches optional
rcParams['figure.figsize'] = 16, 10
# path to images
plot1 = os.path.join(demo_output_directory,"basicTestEnso/ENSO_tel/cmip5_historical_ENSO_tel_ACCESS1-0_r1i1p1_EnsoAmpl_diagnostic_divedown01.png")
plot2 = os.path.join(demo_output_directory,"basicTestEnso/ENSO_tel/cmip5_historical_ENSO_tel_ACCESS1-0_r1i1p1_EnsoAmpl_diagnostic_divedown02.png")
plot3 = os.path.join(demo_output_directory,"basicTestEnso/ENSO_tel/cmip5_historical_ENSO_tel_ACCESS1-0_r1i1p1_EnsoAmpl_diagnostic_divedown03.png")
# display images
fig, ax = plt.subplots(1,3); ax[0].axis('off'); ax[1].axis('off'); ax[2].axis('off')
ax[0].imshow(mpimg.imread(plot1))
ax[1].imshow(mpimg.imread(plot2))
ax[2].imshow(mpimg.imread(plot3))
[13]:
<matplotlib.image.AxesImage at 0x1b050e5c0>
Finally, this example runs the remaining metrics collection ENSO_proc:
[14]:
%%bash -s "$demo_output_directory"
enso_driver.py -p basic_enso_param.py \
--metricsCollection ENSO_proc \
--results_dir $1/basicTestEnso/ENSO_proc
mip: cmip5
exp: historical
models: ['ACCESS1-0']
realization: r1i1p1
mc_name: ENSO_proc
outdir: demo_output/basicTestEnso/ENSO_proc
netcdf_path: demo_output/basicTestEnso/ENSO_proc
debug: False
obs_cmor: True
obs_cmor_path: demo_data/obs4MIPs_PCMDI_monthly
egg_pth: /Users/lee1043/miniforge3/envs/pmp_devel_20250715/share/pmp
output directory for graphics:demo_output/basicTestEnso/ENSO_proc
output directory for diagnostic_results:demo_output/basicTestEnso/ENSO_proc
output directory for metrics_results:demo_output/basicTestEnso/ENSO_proc
list_variables: ['ssh', 'sst', 'taux', 'thf']
list_obs: ['AVISO-1-0', 'ERA-INT', 'GPCP-2-3', 'HadISST-1-1']
Observation dataset AVISO-1-0 is not given for variable thf
Observation dataset GPCP-2-3 is not given for variable thf
Observation dataset HadISST-1-1 is not given for variable thf
PMPdriver: dict_obs readin end
Process start:Fri Aug 29 15:24:07 2025
models: ['ACCESS1-0']
----- model: ACCESS1-0 ---------------------
PMPdriver: var loop start for model ACCESS1-0
realization: r1i1p1
--- run: r1i1p1 ---
--- var: ssh ---
var_in_file: zos
var, areacell_in_file, realm: ssh areacello ocean
path: demo_data/CMIP5_demo_data/zos_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc
file_list: []
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
PMPdriver: var loop end
--- var: sst ---
var_in_file: ts
var, areacell_in_file, realm: sst areacella atmos
path: demo_data/CMIP5_demo_data/ts_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc
file_list: ['demo_data/CMIP5_demo_data/ts_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
PMPdriver: var loop end
--- var: taux ---
var_in_file: tauu
var, areacell_in_file, realm: taux areacella atmos
path: demo_data/CMIP5_demo_data/tauu_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc
file_list: ['demo_data/CMIP5_demo_data/tauu_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
PMPdriver: var loop end
--- var: thf ---
var_in_file: ['hfls', 'hfss', 'rlds', 'rlus', 'rsds', 'rsus']
var, areacell_in_file, realm: thf areacella atmos
path: demo_data/CMIP5_demo_data/hfls_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc
file_list: ['demo_data/CMIP5_demo_data/hfls_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
path: demo_data/CMIP5_demo_data/hfls_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc
file_list: ['demo_data/CMIP5_demo_data/hfls_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
file_areacell_tmp: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
path: demo_data/CMIP5_demo_data/hfss_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc
file_list: ['demo_data/CMIP5_demo_data/hfss_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
file_areacell_tmp: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
path: demo_data/CMIP5_demo_data/rlds_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc
file_list: ['demo_data/CMIP5_demo_data/rlds_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
file_areacell_tmp: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
path: demo_data/CMIP5_demo_data/rlus_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc
file_list: ['demo_data/CMIP5_demo_data/rlus_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
file_areacell_tmp: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
path: demo_data/CMIP5_demo_data/rsds_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc
file_list: ['demo_data/CMIP5_demo_data/rsds_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
file_areacell_tmp: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
path: demo_data/CMIP5_demo_data/rsus_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc
file_list: ['demo_data/CMIP5_demo_data/rsus_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc']
path: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
file_list: ['demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc']
file_areacell_tmp: demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc
PMPdriver: var loop end
dictDatasets:
{
"model": {
"ACCESS1-0_r1i1p1": {
"ssh": {
"areaname": "areacello",
"landmaskname": "sftlf",
"path + filename": null,
"path + filename_area": "demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"path + filename_landmask": null,
"varname": "zos"
},
"sst": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/CMIP5_demo_data/ts_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc",
"path + filename_area": "demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"path + filename_landmask": "demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"varname": "ts"
},
"taux": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/CMIP5_demo_data/tauu_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc",
"path + filename_area": "demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"path + filename_landmask": "demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"varname": "tauu"
},
"thf": {
"areaname": [
"areacella",
"areacella",
"areacella",
"areacella",
"areacella",
"areacella"
],
"landmaskname": [
"sftlf",
"sftlf",
"sftlf",
"sftlf",
"sftlf",
"sftlf"
],
"path + filename": [
"demo_data/CMIP5_demo_data/hfls_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc",
"demo_data/CMIP5_demo_data/hfss_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc",
"demo_data/CMIP5_demo_data/rlds_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc",
"demo_data/CMIP5_demo_data/rlus_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc",
"demo_data/CMIP5_demo_data/rsds_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc",
"demo_data/CMIP5_demo_data/rsus_Amon_ACCESS1-0_historical_r1i1p1_185001-200512.nc"
],
"path + filename_area": [
"demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc"
],
"path + filename_landmask": [
"demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc",
"demo_data/CMIP5_demo_data/sftlf_fx_ACCESS1-0_amip_r0i0p0.nc"
],
"varname": [
"hfls",
"hfss",
"rlds",
"rlus",
"rsds",
"rsus"
]
}
}
},
"observations": {
"AVISO": {
"ssh": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/obs4MIPs_PCMDI_monthly/CNES/AVISO-1-0/mon/zos/gn/v20210727/zos_mon_AVISO-1-0_PCMDI_gn_199301-201912.nc",
"path + filename_area": null,
"path + filename_landmask": null,
"varname": "zos"
}
},
"ERA-Interim": {
"sst": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/obs4MIPs_PCMDI_monthly/ECMWF/ERA-INT/mon/ts/gn/v20210727/ts_mon_ERA-INT_PCMDI_gn_197901-201903.nc",
"path + filename_area": null,
"path + filename_landmask": null,
"varname": "ts"
},
"taux": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/obs4MIPs_PCMDI_monthly/ECMWF/ERA-INT/mon/tauu/gn/v20210727/tauu_mon_ERA-INT_PCMDI_gn_197901-201903.nc",
"path + filename_area": null,
"path + filename_landmask": null,
"varname": "tauu"
},
"thf": {
"areaname": [
"areacella",
"areacella",
"areacella",
"areacella",
"areacella",
"areacella"
],
"landmaskname": [
"sftlf",
"sftlf",
"sftlf",
"sftlf",
"sftlf",
"sftlf"
],
"path + filename": [
"demo_data/obs4MIPs_PCMDI_monthly/ECMWF/ERA-INT/mon/hfls/gn/v20210727/hfls_mon_ERA-INT_PCMDI_gn_197901-201903.nc",
"demo_data/obs4MIPs_PCMDI_monthly/ECMWF/ERA-INT/mon/hfss/gn/v20210727/hfss_mon_ERA-INT_PCMDI_gn_197901-201903.nc",
"demo_data/obs4MIPs_PCMDI_monthly/ECMWF/ERA-INT/mon/rlds/gn/v20210727/rlds_mon_ERA-INT_PCMDI_gn_197901-201903.nc",
"demo_data/obs4MIPs_PCMDI_monthly/ECMWF/ERA-INT/mon/rlus/gn/v20210727/rlus_mon_ERA-INT_PCMDI_gn_197901-201903.nc",
"demo_data/obs4MIPs_PCMDI_monthly/ECMWF/ERA-INT/mon/rsds/gn/v20210727/rsds_mon_ERA-INT_PCMDI_gn_197901-201903.nc",
"demo_data/obs4MIPs_PCMDI_monthly/ECMWF/ERA-INT/mon/rsus/gn/v20210727/rsus_mon_ERA-INT_PCMDI_gn_197901-201903.nc"
],
"path + filename_area": [
null,
null,
null,
null,
null,
null
],
"path + filename_landmask": null,
"varname": [
"hfls",
"hfss",
"rlds",
"rlus",
"rsds",
"rsus"
]
}
},
"HadISST": {
"sst": {
"areaname": "areacella",
"landmaskname": "sftlf",
"path + filename": "demo_data/obs4MIPs_PCMDI_monthly/MOHC/HadISST-1-1/mon/ts/gn/v20210727/ts_mon_HadISST-1-1_PCMDI_gn_187001-201907.nc",
"path + filename_area": null,
"path + filename_landmask": null,
"varname": "ts"
}
}
}
}
### Compute the metric collection ###
ComputeCollection: metric = BiasSstLonRmse
ComputeMetric: oneVarRMSmetric, BiasSstLonRmse = ACCESS1-0_r1i1p1 and ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = BiasTauxLonRmse
ComputeMetric: oneVarRMSmetric, BiasTauxLonRmse = ACCESS1-0_r1i1p1 and ERA-Interim
EnsoUvcdatToolsLib AverageHorizontal
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoAmpl
ComputeMetric: oneVarmetric = ACCESS1-0_r1i1p1
EnsoUvcdatToolsLib AverageMeridional
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/cdms2/dataset.py:2179: Warning: Files are written with compression and no shuffling
You can query different values of compression using the functions:
cdms2.getNetcdfShuffleFlag() returning 1 if shuffling is enabled, 0 otherwise
cdms2.getNetcdfDeflateFlag() returning 1 if deflate is used, 0 otherwise
cdms2.getNetcdfDeflateLevelFlag() returning the level of compression for the deflate method
If you want to turn that off or set different values of compression use the functions:
value = 0
cdms2.setNetcdfShuffleFlag(value) ## where value is either 0 or 1
cdms2.setNetcdfDeflateFlag(value) ## where value is either 0 or 1
cdms2.setNetcdfDeflateLevelFlag(value) ## where value is a integer between 0 and 9 included
To produce NetCDF3 Classic files use:
cdms2.useNetCDF3()
To Force NetCDF4 output with classic format and no compressing use:
cdms2.setNetcdf4Flag(1)
NetCDF4 file with no shuffling or deflate and noclassic will be open for parallel i/o
warnings.warn("Files are written with compression and no shuffling\n" +
ComputeMetric: oneVarmetric = ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsodSstOce_2
ComputeMetric: twoVarmetric = ACCESS1-0_r1i1p1
EnsoUvcdatToolsLib ReadAndSelectRegion
hfls sign reversed
range old = +1.99 to +243.32
range new = -243.32 to -1.99
EnsoUvcdatToolsLib ReadAndSelectRegion
hfss sign reversed
range old = -3.28 to +31.07
range new = -31.07 to +3.28
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/cdms2/MV2.py:318: Warning: arguments order for compress function has changed
it is now: MV2.copmress(array,condition), if your code seems to not react or act wrong to a call to compress, please check this
warnings.warn(
EnsoUvcdatToolsLib ReadAndSelectRegion
hfls sign reversed
range old = +1.99 to +290.82
range new = -290.82 to -1.99
EnsoUvcdatToolsLib ReadAndSelectRegion
hfss sign reversed
range old = -3.28 to +62.31
range new = -62.31 to +3.28
EnsoUvcdatToolsLib AverageMeridional
EnsoUvcdatToolsLib AverageMeridional
ComputeMetric: twoVarmetric = ERA-Interim_ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoFbSshSst
ComputeCollection: ENSO_proc, metric EnsoFbSshSst not computed
reason(s):
no modeled ssh given
ComputeCollection: metric = EnsoFbSstTaux
ComputeMetric: twoVarmetric = ACCESS1-0_r1i1p1
EnsoUvcdatToolsLib AverageHorizontal
EnsoUvcdatToolsLib AverageHorizontal
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoFbSstThf
ComputeMetric: twoVarmetric = ACCESS1-0_r1i1p1
EnsoUvcdatToolsLib ReadAndSelectRegion
hfls sign reversed
range old = +1.99 to +243.32
range new = -243.32 to -1.99
EnsoUvcdatToolsLib ReadAndSelectRegion
hfss sign reversed
range old = -3.28 to +31.07
range new = -31.07 to +3.28
EnsoUvcdatToolsLib ReadAndSelectRegion
hfls sign reversed
range old = +1.99 to +290.82
range new = -290.82 to -1.99
EnsoUvcdatToolsLib ReadAndSelectRegion
hfss sign reversed
range old = -3.28 to +62.31
range new = -62.31 to +3.28
EnsoUvcdatToolsLib AverageMeridional
EnsoUvcdatToolsLib AverageMeridional
ComputeMetric: twoVarmetric = ERA-Interim_ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoFbTauxSsh
ComputeCollection: ENSO_proc, metric EnsoFbTauxSsh not computed
reason(s):
no modeled ssh given
ComputeCollection: metric = EnsoSeasonality
ComputeMetric: oneVarmetric = ACCESS1-0_r1i1p1
EnsoUvcdatToolsLib AverageMeridional
EnsoUvcdatToolsLib AverageMeridional
EnsoUvcdatToolsLib AverageMeridional
ComputeMetric: oneVarmetric = ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoSstLonRmse
ComputeMetric: oneVarRMSmetric, EnsoSstLonRmse = ACCESS1-0_r1i1p1 and ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
ComputeCollection: metric = EnsoSstSkew
ComputeMetric: oneVarmetric = ACCESS1-0_r1i1p1
EnsoUvcdatToolsLib AverageMeridional
ComputeMetric: oneVarmetric = ERA-Interim
NOTE: Estimated landmask applied
/Users/lee1043/miniforge3/envs/pmp_devel_20250715/lib/python3.10/site-packages/share/cdutil/navy_land.nc
INFO::2025-08-29 15:26::pcmdi_metrics:: Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_proc/cmip5_historical_ENSO_proc_basicTestEnso_ACCESS1-0_r1i1p1.json
2025-08-29 15:26:11,380 [INFO]: base.py(write:347) >> Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_proc/cmip5_historical_ENSO_proc_basicTestEnso_ACCESS1-0_r1i1p1.json
2025-08-29 15:26:11,380 [INFO]: base.py(write:347) >> Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_proc/cmip5_historical_ENSO_proc_basicTestEnso_ACCESS1-0_r1i1p1.json
INFO::2025-08-29 15:26::pcmdi_metrics:: Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_proc/cmip5_historical_ENSO_proc_basicTestEnso_ACCESS1-0_r1i1p1_diveDown.json
2025-08-29 15:26:16,871 [INFO]: base.py(write:347) >> Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_proc/cmip5_historical_ENSO_proc_basicTestEnso_ACCESS1-0_r1i1p1_diveDown.json
2025-08-29 15:26:16,871 [INFO]: base.py(write:347) >> Results saved to a json file: /Users/lee1043/Documents/Research/git/pcmdi_metrics_20230620_pcmdi/pcmdi_metrics/doc/jupyter/Demo/demo_output/basicTestEnso/ENSO_proc/cmip5_historical_ENSO_proc_basicTestEnso_ACCESS1-0_r1i1p1_diveDown.json
figure plotting start
metrics: ['BiasSstLonRmse', 'BiasTauxLonRmse', 'EnsoAmpl', 'EnsodSstOce_2', 'EnsoFbSshSst', 'EnsoFbSstTaux', 'EnsoFbSstThf', 'EnsoFbTauxSsh', 'EnsoSeasonality', 'EnsoSstLonRmse', 'EnsoSstSkew']
filename_js: demo_output/basicTestEnso/ENSO_proc/cmip5_historical_ENSO_proc_basicTestEnso_ACCESS1-0_r1i1p1.json
met: BiasSstLonRmse
filename_nc: demo_output/basicTestEnso/ENSO_proc/cmip5_historical_ENSO_proc_basicTestEnso_ACCESS1-0_r1i1p1_BiasSstLonRmse.nc
failed for ACCESS1-0 r1i1p1
'BiasSstLonRmse'
PMPdriver: model loop end
Process end: Fri Aug 29 15:26:16 2025
[15]:
# figure size in inches optional
rcParams['figure.figsize'] = 16, 10
# path to images
plot1 = os.path.join(demo_output_directory,"basicTestEnso/ENSO_proc/cmip5_historical_ENSO_proc_ACCESS1-0_r1i1p1_EnsoSstSkew_diagnostic_divedown01.png")
plot2 = os.path.join(demo_output_directory,"basicTestEnso/ENSO_proc/cmip5_historical_ENSO_proc_ACCESS1-0_r1i1p1_EnsoSstSkew_diagnostic_divedown02.png")
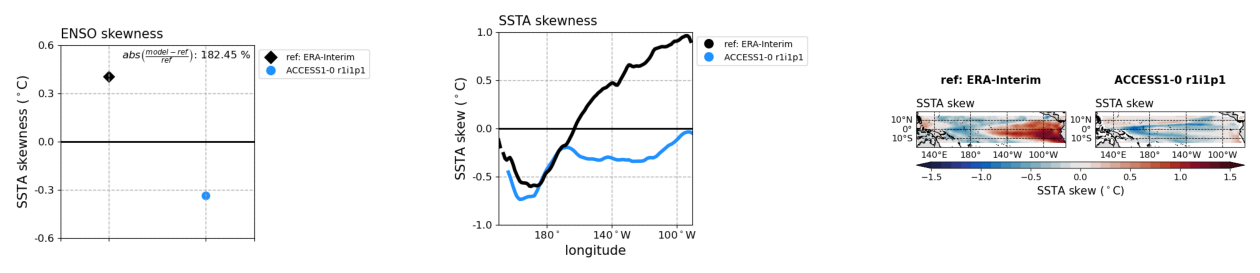
plot3 = os.path.join(demo_output_directory,"basicTestEnso/ENSO_proc/cmip5_historical_ENSO_proc_ACCESS1-0_r1i1p1_EnsoSstSkew_diagnostic_divedown03.png")
# display images
fig, ax = plt.subplots(1,3); ax[0].axis('off'); ax[1].axis('off'); ax[2].axis('off')
ax[0].imshow(mpimg.imread(plot1))
ax[1].imshow(mpimg.imread(plot2))
ax[2].imshow(mpimg.imread(plot3))
[15]:
<matplotlib.image.AxesImage at 0x1b8e107c0>