PMP mean climate: Compare user’s model with CMIP6
In this document, you will visualize PMP’s mean climate results for your own model, together with CMIP6, to compare performace. You will create following plots.
Portrait plot
Parallel coordinate plot
Box plot
Taylor Diagram
Written by Jiwoo Lee (LLNL/PCMDI), with contribution from Ana Ordonez (LLNL/PCMDI) and John Krasting (NOAA/GFDL)
Last update: September 2022
Contents
Read data from JSON files
1.1 Download PMP output JSON files for CMIP models
1.2 Provide PMP results for the user’s model
1.3 Extract data from JSON files
1.4 Combine your PMP output with CMIP results
1.5 Normalize each column by its median for portrait plot
Portrait Plot
Parallel Coordinate Plot
Box plots
Taylor Diagram
5.1 Identify all models
5.2 Highlight user’s model
1. Read data from JSON files
Input data for portrait plot is expected as a set a (stacked or list of) 2-d numpy array(s) with list of strings for x and y axes labels.
1.1 Download PMP output JSON files for CMIP models
[1]:
import glob
import os
import numpy as np
from pcmdi_metrics.graphics import download_archived_results
PMP output files downloadable from the PMP results archive.
[2]:
vars = ['pr', 'prw', 'psl', 'rlds', 'rltcre', 'rlus', 'rlut', 'rlutcs', 'rsds', 'rsdscs', 'rsdt', 'rstcre', 'rsut', 'rsutcs', 'sfcWind',
'ta-200', 'ta-850', 'tas', 'tauu', 'ts', 'ua-200', 'ua-850', 'va-200', 'va-850', 'zg-500']
mip = "cmip6"
exp = "historical"
data_version = "v20210811"
json_dir = './json_files'
Provide directory path and filename in the PMP results archive.
[3]:
for var in vars:
path = "metrics_results/mean_climate/"+mip+"/"+exp+"/"+data_version+"/"+var+"."+mip+"."+exp+".regrid2.2p5x2p5."+data_version+".json"
download_archived_results(path, json_dir)
Check JSON files
[4]:
json_list = sorted(glob.glob(os.path.join(json_dir, '*' + mip + '*' + data_version + '.json')))
for json_file in json_list:
print(json_file.split('/')[-1])
pr.cmip6.historical.regrid2.2p5x2p5.v20210811.json
prw.cmip6.historical.regrid2.2p5x2p5.v20210811.json
psl.cmip6.historical.regrid2.2p5x2p5.v20210811.json
rlds.cmip6.historical.regrid2.2p5x2p5.v20210811.json
rltcre.cmip6.historical.regrid2.2p5x2p5.v20210811.json
rlus.cmip6.historical.regrid2.2p5x2p5.v20210811.json
rlut.cmip6.historical.regrid2.2p5x2p5.v20210811.json
rlutcs.cmip6.historical.regrid2.2p5x2p5.v20210811.json
rsds.cmip6.historical.regrid2.2p5x2p5.v20210811.json
rsdscs.cmip6.historical.regrid2.2p5x2p5.v20210811.json
rsdt.cmip6.historical.regrid2.2p5x2p5.v20210811.json
rstcre.cmip6.historical.regrid2.2p5x2p5.v20210811.json
rsut.cmip6.historical.regrid2.2p5x2p5.v20210811.json
rsutcs.cmip6.historical.regrid2.2p5x2p5.v20210811.json
sfcWind.cmip6.historical.regrid2.2p5x2p5.v20210811.json
ta-200.cmip6.historical.regrid2.2p5x2p5.v20210811.json
ta-850.cmip6.historical.regrid2.2p5x2p5.v20210811.json
tas.cmip6.historical.regrid2.2p5x2p5.v20210811.json
tauu.cmip6.historical.regrid2.2p5x2p5.v20210811.json
ts.cmip6.historical.regrid2.2p5x2p5.v20210811.json
ua-200.cmip6.historical.regrid2.2p5x2p5.v20210811.json
ua-850.cmip6.historical.regrid2.2p5x2p5.v20210811.json
va-200.cmip6.historical.regrid2.2p5x2p5.v20210811.json
va-850.cmip6.historical.regrid2.2p5x2p5.v20210811.json
zg-500.cmip6.historical.regrid2.2p5x2p5.v20210811.json
1.2 Provide PMP results for the user’s model
[5]:
json_dir_test = './json_files_test_case'
[6]:
for var in vars:
path = "test_case/mean_climate/my_model."+var+".regrid2.2p5x2p5.json"
download_archived_results(path, json_dir_test)
test_case/mean_climate/my_model.prw.regrid2.2p5x2p5.json not exist in https://github.com/PCMDI/pcmdi_metrics_results_archive/tree/main/
test_case/mean_climate/my_model.rsdscs.regrid2.2p5x2p5.json not exist in https://github.com/PCMDI/pcmdi_metrics_results_archive/tree/main/
test_case/mean_climate/my_model.tauu.regrid2.2p5x2p5.json not exist in https://github.com/PCMDI/pcmdi_metrics_results_archive/tree/main/
[7]:
json_list_test = sorted(glob.glob(os.path.join(json_dir_test, '*.json')))
for json_file in json_list_test:
print(json_file.split('/')[-1])
my_model.pr.regrid2.2p5x2p5.json
my_model.psl.regrid2.2p5x2p5.json
my_model.rlds.regrid2.2p5x2p5.json
my_model.rltcre.regrid2.2p5x2p5.json
my_model.rlus.regrid2.2p5x2p5.json
my_model.rlut.regrid2.2p5x2p5.json
my_model.rlutcs.regrid2.2p5x2p5.json
my_model.rsds.regrid2.2p5x2p5.json
my_model.rsdt.regrid2.2p5x2p5.json
my_model.rstcre.regrid2.2p5x2p5.json
my_model.rsut.regrid2.2p5x2p5.json
my_model.rsutcs.regrid2.2p5x2p5.json
my_model.sfcWind.regrid2.2p5x2p5.json
my_model.ta-200.regrid2.2p5x2p5.json
my_model.ta-850.regrid2.2p5x2p5.json
my_model.tas.regrid2.2p5x2p5.json
my_model.ts.regrid2.2p5x2p5.json
my_model.ua-200.regrid2.2p5x2p5.json
my_model.ua-850.regrid2.2p5x2p5.json
my_model.va-200.regrid2.2p5x2p5.json
my_model.va-850.regrid2.2p5x2p5.json
my_model.zg-500.regrid2.2p5x2p5.json
1.3 Extract data from JSON files
Use Metrics class (that use read_mean_clim_json_files function underneath) to extract data from the above JSON files.
Parameters
json_list: list of string, where each element is for path/file for PMP output JSON files
Returned object includes
df_dict: dictionary that has[stat][season][region]hierarchy structure storing pandas dataframe for metric numbers (Rows: models, Columns: variables (i.e., 2d array)var_list: list of string, all variables from JSON filesvar_unit_list: list of string, all variables and its units from JSON filesvar_ref_dict: dictonary for reference dataset used for each variableregions: list of string, regionsstats: list of string, statistics
[8]:
from pcmdi_metrics.graphics import Metrics
[9]:
# existing library of mean climate metrics downloaded
library = Metrics(json_list)
Warning: The provided level value 20000 appears to be in Pa. It will be automatically converted to hPa by dividing by 100.
Warning: The provided level value 85000 appears to be in Pa. It will be automatically converted to hPa by dividing by 100.
Warning: The provided level value 20000 appears to be in Pa. It will be automatically converted to hPa by dividing by 100.
Warning: The provided level value 85000 appears to be in Pa. It will be automatically converted to hPa by dividing by 100.
Warning: The provided level value 20000 appears to be in Pa. It will be automatically converted to hPa by dividing by 100.
Warning: The provided level value 85000 appears to be in Pa. It will be automatically converted to hPa by dividing by 100.
Warning: The provided level value 50000 appears to be in Pa. It will be automatically converted to hPa by dividing by 100.
[10]:
# new test case
test_case = Metrics(json_list_test)
Warning: The provided level value 20000 appears to be in Pa. It will be automatically converted to hPa by dividing by 100.
Warning: The provided level value 85000 appears to be in Pa. It will be automatically converted to hPa by dividing by 100.
Warning: The provided level value 20000 appears to be in Pa. It will be automatically converted to hPa by dividing by 100.
Warning: The provided level value 85000 appears to be in Pa. It will be automatically converted to hPa by dividing by 100.
Warning: The provided level value 20000 appears to be in Pa. It will be automatically converted to hPa by dividing by 100.
Warning: The provided level value 85000 appears to be in Pa. It will be automatically converted to hPa by dividing by 100.
Warning: The provided level value 50000 appears to be in Pa. It will be automatically converted to hPa by dividing by 100.
1.4 Combine your PMP output with CMIP results
[11]:
# merged
combined = library.merge(test_case)
[12]:
df_dict = combined.df_dict
var_list = combined.var_list
var_unit_list = combined.var_unit_list
var_ref_dict = combined.var_ref_dict
regions = combined.regions
stats = combined.stats
Check loaded data
[13]:
print('var_list:', sorted(var_list))
print('var_unit_list:', sorted(var_unit_list))
print('var_ref_dict:', var_ref_dict)
var_list: ['pr', 'prw', 'psl', 'rlds', 'rltcre', 'rlus', 'rlut', 'rlutcs', 'rsds', 'rsdscs', 'rsdt', 'rstcre', 'rsut', 'rsutcs', 'sfcWind', 'ta-200', 'ta-850', 'tas', 'tauu', 'ts', 'ua-200', 'ua-850', 'va-200', 'va-850', 'zg-500']
var_unit_list: ['pr [N/A]', 'pr [kg m-2 s-1]', 'prw [N/A]', 'psl [N/A]', 'psl [Pa]', 'rlds [N/A]', 'rlds [W m-2]', 'rltcre [W m-2]', 'rlus [N/A]', 'rlus [W m-2]', 'rlut [N/A]', 'rlut [W m-2]', 'rlutcs [N/A]', 'rlutcs [W m-2]', 'rsds [N/A]', 'rsds [W m-2]', 'rsdscs [N/A]', 'rsdt [N/A]', 'rsdt [W m-2]', 'rstcre [W m-2]', 'rsut [N/A]', 'rsut [W m-2]', 'rsutcs [N/A]', 'rsutcs [W m-2]', 'sfcWind [N/A]', 'sfcWind [m s-1]', 'ta-200 [K]', 'ta-200 [N/A]', 'ta-850 [K]', 'ta-850 [N/A]', 'tas [K]', 'tas [N/A]', 'tauu [N/A]', 'ts [K]', 'ts [N/A]', 'ua-200 [N/A]', 'ua-200 [m s-1]', 'ua-850 [N/A]', 'ua-850 [m s-1]', 'va-200 [N/A]', 'va-200 [m s-1]', 'va-850 [N/A]', 'va-850 [m s-1]', 'zg-500 [N/A]', 'zg-500 [m]']
var_ref_dict: {'pr': 'GPCP-2-3', 'prw': 'REMSS-PRW-v07r01', 'psl': 'ERA-5', 'rlds': 'CERES-EBAF-4-1', 'rltcre': 'CERES-EBAF-4-1', 'rlus': 'CERES-EBAF-4-1', 'rlut': 'CERES-EBAF-4-1', 'rlutcs': 'CERES-EBAF-4-1', 'rsds': 'CERES-EBAF-4-1', 'rsdscs': 'CERES-EBAF-4-1', 'rsdt': 'CERES-EBAF-4-1', 'rstcre': 'CERES-EBAF-4-1', 'rsut': 'CERES-EBAF-4-1', 'rsutcs': 'CERES-EBAF-4-1', 'sfcWind': 'REMSS-PRW-v07r01', 'ta-200': 'ERA-5', 'ta-850': 'ERA-5', 'tas': 'ERA-5', 'tauu': 'ERA-INT', 'ts': 'ERA-5', 'ua-200': 'ERA-5', 'ua-850': 'ERA-5', 'va-200': 'ERA-5', 'va-850': 'ERA-5', 'zg-500': 'ERA-5'}
In pandas dataframe, you will see your model is now included in the last row, as below.
[14]:
df_dict['rms_xy']['djf']['global']
[14]:
| model | run | model_run | pr | prw | psl | rlds | rltcre | rlus | rlut | ... | ta-200 | ta-850 | tas | tauu | ts | ua-200 | ua-850 | va-200 | va-850 | zg-500 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ACCESS-CM2 | r1i1p1 | ACCESS-CM2_r1i1p1 | 1.661 | 139.039 | 245.952 | 11.552 | 8.519 | 10.560 | 10.991 | ... | 2.717 | 1.495 | 2.508 | 0.040 | 2.770 | 4.174 | 1.421 | 1.689 | 0.799 | 26.053 |
| 1 | ACCESS-ESM1-5 | r1i1p1 | ACCESS-ESM1-5_r1i1p1 | 1.739 | 139.068 | 215.795 | 10.553 | 7.358 | 10.352 | 10.875 | ... | 2.551 | 1.335 | 2.106 | 0.035 | 2.250 | 3.305 | 1.408 | 1.804 | 0.859 | 25.470 |
| 2 | AWI-CM-1-1-MR | r1i1p1 | AWI-CM-1-1-MR_r1i1p1 | 1.604 | 139.179 | 170.936 | 11.231 | 7.957 | 8.496 | 8.960 | ... | 1.847 | 1.074 | 1.414 | 0.028 | 1.609 | 2.483 | 1.251 | 1.586 | 0.711 | 16.738 |
| 3 | AWI-ESM-1-1-LR | r1i1p1 | AWI-ESM-1-1-LR_r1i1p1 | 1.937 | 139.199 | 201.589 | 14.743 | 8.935 | 12.495 | 11.630 | ... | 3.966 | 2.224 | 2.039 | 0.034 | 2.317 | 3.835 | 1.684 | 2.241 | 1.024 | NaN |
| 4 | BCC-CSM2-MR | r1i1p1 | BCC-CSM2-MR_r1i1p1 | 1.700 | 139.251 | 237.180 | 13.025 | 7.370 | 11.014 | 9.984 | ... | NaN | NaN | 2.669 | 0.034 | 2.585 | NaN | NaN | NaN | NaN | NaN |
| 5 | BCC-ESM1 | r1i1p1 | BCC-ESM1_r1i1p1 | 1.631 | 139.152 | 237.973 | 14.469 | 8.423 | 12.970 | 12.763 | ... | 4.343 | 2.055 | 3.211 | 0.038 | 3.188 | 4.673 | 1.849 | 2.119 | 1.118 | NaN |
| 6 | CAMS-CSM1-0 | r1i1p1 | CAMS-CSM1-0_r1i1p1 | 1.721 | 139.513 | 182.945 | 19.644 | 8.397 | 14.631 | 11.507 | ... | NaN | NaN | 3.462 | NaN | 3.858 | NaN | NaN | NaN | NaN | NaN |
| 7 | CanESM5 | r1i1p1 | CanESM5_r1i1p1 | 1.779 | 138.924 | 240.049 | 16.095 | 8.406 | 12.479 | 11.047 | ... | 3.066 | 1.905 | 2.465 | 0.055 | 2.622 | 4.055 | 1.706 | 2.013 | 1.008 | 32.574 |
| 8 | CESM2 | r1i1p1 | CESM2_r1i1p1 | 1.234 | 139.212 | 210.704 | 11.026 | 6.979 | 9.611 | 7.908 | ... | 2.057 | 1.309 | 1.483 | 0.089 | 1.800 | 3.433 | 1.377 | 1.973 | 0.800 | NaN |
| 9 | CESM2-FV2 | r1i1p1 | CESM2-FV2_r1i1p1 | 1.397 | 139.161 | 242.138 | 12.376 | 7.835 | 10.690 | 8.740 | ... | 3.026 | 1.987 | 1.892 | 0.090 | 2.142 | 4.017 | 2.061 | 2.204 | 0.988 | NaN |
| 10 | CESM2-WACCM | r1i1p1 | CESM2-WACCM_r1i1p1 | 1.203 | 139.166 | 205.141 | 10.544 | 6.761 | 9.166 | 7.680 | ... | NaN | NaN | 1.462 | 0.088 | 1.750 | 3.131 | 1.481 | 1.761 | 0.787 | NaN |
| 11 | CESM2-WACCM-FV2 | r1i1p1 | CESM2-WACCM-FV2_r1i1p1 | 1.517 | 139.211 | 194.140 | 12.047 | 7.890 | 10.551 | 9.264 | ... | NaN | NaN | 1.744 | 0.089 | 2.095 | 3.086 | 1.871 | 1.976 | 0.903 | 22.056 |
| 12 | CIESM | r1i1p1 | CIESM_r1i1p1 | 3.627 | 139.554 | 175.697 | 10.636 | 8.130 | 8.867 | 7.649 | ... | 3.773 | 1.221 | 1.512 | 0.151 | 1.802 | 3.106 | 1.264 | 1.635 | 0.834 | 39.275 |
| 13 | CMCC-CM2-HR4 | r1i1p1 | CMCC-CM2-HR4_r1i1p1 | 1.599 | 139.313 | 327.559 | 13.640 | 8.576 | 9.807 | 10.873 | ... | 3.350 | 1.575 | 1.675 | 0.175 | 1.865 | 4.592 | 2.092 | 2.294 | 1.140 | 41.643 |
| 14 | CMCC-CM2-SR5 | r1i1p1 | CMCC-CM2-SR5_r1i1p1 | 1.529 | 139.410 | 294.997 | 11.556 | 8.437 | 10.053 | 8.629 | ... | 3.479 | 1.774 | 1.903 | 0.150 | 2.216 | 3.781 | 1.530 | 1.770 | 0.886 | 29.599 |
| 15 | E3SM-1-0 | r1i1p1 | E3SM-1-0_r1i1p1 | 1.358 | 139.355 | 263.067 | 13.937 | 6.388 | 10.892 | 7.968 | ... | 2.954 | 2.322 | 2.156 | 0.048 | 2.312 | 3.283 | 1.538 | 1.759 | 0.846 | 31.321 |
| 16 | E3SM-1-1 | r1i1p1 | E3SM-1-1_r1i1p1 | 1.384 | 139.305 | 286.393 | 14.353 | 6.715 | 11.588 | 8.222 | ... | 3.055 | NaN | 2.293 | 0.037 | 2.429 | 3.477 | NaN | 1.954 | NaN | 33.610 |
| 17 | E3SM-1-1-ECA | r1i1p1 | E3SM-1-1-ECA_r1i1p1 | 1.349 | 139.318 | 277.138 | 14.140 | 6.519 | 11.746 | 8.159 | ... | 3.094 | 26.727 | 2.367 | 0.037 | 2.507 | 3.483 | 1.555 | 1.871 | 0.885 | 36.308 |
| 18 | EC-Earth3 | r1i1p1 | EC-Earth3_r1i1p1 | 1.239 | 139.257 | 167.478 | 15.160 | 6.863 | 12.768 | 9.741 | ... | 1.701 | 1.755 | 2.450 | 0.024 | 3.674 | 3.163 | 0.991 | 1.662 | 0.677 | 31.607 |
| 19 | EC-Earth3-AerChem | r1i1p1 | EC-Earth3-AerChem_r1i1p1 | 1.291 | 139.269 | 176.365 | 14.411 | 6.825 | 12.037 | 9.629 | ... | NaN | NaN | 2.229 | 0.025 | 3.634 | 3.352 | 1.033 | 1.599 | 0.669 | 33.658 |
| 20 | EC-Earth3-Veg | r1i1p1 | EC-Earth3-Veg_r1i1p1 | 1.262 | 139.308 | 159.077 | 13.688 | 6.945 | 11.592 | 9.661 | ... | 1.613 | 1.635 | 2.004 | 0.026 | 3.610 | 3.315 | 1.015 | 1.534 | 0.677 | 28.996 |
| 21 | EC-Earth3-Veg-LR | r1i1p1 | EC-Earth3-Veg-LR_r1i1p1 | 1.266 | 139.286 | 158.686 | 16.394 | 6.922 | 13.260 | 10.065 | ... | 2.019 | 1.797 | 2.502 | 0.024 | 3.708 | 3.264 | 1.098 | 1.591 | 0.706 | 33.999 |
| 22 | FGOALS-f3-L | r1i1p1 | FGOALS-f3-L_r1i1p1 | 1.449 | NaN | 359.037 | 11.759 | 10.375 | 12.101 | 10.129 | ... | 3.215 | 1.683 | 2.779 | 0.171 | 2.889 | 4.494 | 1.693 | 2.272 | 0.973 | 54.113 |
| 23 | FGOALS-g3 | r1i1p1 | FGOALS-g3_r1i1p1 | 1.658 | 138.888 | 292.081 | 19.393 | 9.125 | 15.059 | 12.801 | ... | 5.826 | 1.823 | 3.562 | 0.163 | 3.895 | 3.641 | 1.909 | 2.136 | 1.050 | 29.997 |
| 24 | FIO-ESM-2-0 | r1i1p1 | FIO-ESM-2-0_r1i1p1 | 1.388 | NaN | 228.979 | 10.459 | 7.668 | 8.932 | 9.352 | ... | 3.792 | 1.551 | 1.586 | 0.151 | 1.776 | 3.900 | 1.462 | 2.196 | 0.890 | 26.022 |
| 25 | GFDL-CM4 | r1i1p1 | GFDL-CM4_r1i1p1 | 1.337 | 139.040 | 196.965 | 14.900 | 6.849 | 9.760 | 8.148 | ... | 2.109 | 1.830 | 2.331 | 0.027 | 2.226 | 2.844 | 1.379 | 1.323 | 0.707 | 42.543 |
| 26 | GFDL-ESM4 | r1i1p1 | GFDL-ESM4_r1i1p1 | 1.375 | 139.071 | 204.671 | 12.807 | 6.885 | 9.041 | 8.459 | ... | NaN | NaN | 2.034 | 0.028 | 2.067 | 2.754 | 1.376 | 1.457 | 0.730 | NaN |
| 27 | GISS-E2-1-G | r1i1p1 | GISS-E2-1-G_r1i1p1 | 1.631 | 139.025 | 302.630 | 15.092 | 9.973 | 12.631 | 11.219 | ... | 3.685 | 2.058 | 2.643 | 0.047 | 2.867 | 5.480 | 1.832 | 2.889 | 1.128 | 46.220 |
| 28 | GISS-E2-1-G-CC | r1i1p1 | GISS-E2-1-G-CC_r1i1p1 | 1.699 | 139.058 | 302.395 | 15.650 | 10.280 | 12.949 | 11.922 | ... | 3.737 | 2.034 | 2.663 | 0.046 | 2.905 | 5.951 | 1.898 | 3.107 | 1.159 | 45.968 |
| 29 | GISS-E2-1-H | r1i1p1 | GISS-E2-1-H_r1i1p1 | 1.675 | 139.224 | 486.524 | 16.427 | 9.876 | 13.117 | 11.333 | ... | 3.881 | 1.990 | 2.482 | 0.050 | 2.806 | 5.074 | 1.890 | 2.974 | 1.166 | 56.408 |
| 30 | INM-CM4-8 | r1i1p1 | INM-CM4-8_r1i1p1 | 1.724 | 139.201 | 243.852 | 18.399 | 10.049 | 11.774 | 12.460 | ... | 3.799 | 2.989 | 2.597 | 0.074 | 2.769 | 5.865 | 2.035 | 2.760 | 1.199 | 52.334 |
| 31 | INM-CM5-0 | r1i1p1 | INM-CM5-0_r1i1p1 | 1.667 | 139.189 | 197.334 | 16.972 | 8.735 | 10.580 | 10.378 | ... | NaN | NaN | 2.199 | 0.071 | 2.380 | 4.022 | 1.688 | 2.309 | 1.074 | 51.208 |
| 32 | IPSL-CM6A-LR | r1i1p1 | IPSL-CM6A-LR_r1i1p1 | 1.625 | 139.137 | 240.321 | 13.602 | 7.105 | 9.523 | 8.710 | ... | NaN | NaN | 2.073 | 0.036 | 1.987 | 3.033 | 1.661 | 1.803 | 0.973 | 63.596 |
| 33 | KACE-1-0-G | r1i1p1 | KACE-1-0-G_r1i1p1 | 1.457 | 139.082 | 208.175 | 11.662 | 8.174 | 11.210 | 10.097 | ... | 3.300 | 1.399 | 2.462 | 0.036 | 2.715 | 3.707 | 1.346 | 1.845 | 0.825 | 23.343 |
| 34 | MCM-UA-1-0 | r1i1p1 | MCM-UA-1-0_r1i1p1 | 1.605 | NaN | 355.577 | NaN | NaN | NaN | 14.301 | ... | 2.990 | 4.290 | 3.190 | 0.150 | 3.289 | 4.458 | 1.929 | 2.470 | 1.142 | 63.850 |
| 35 | MIROC6 | r1i1p1 | MIROC6_r1i1p1 | 1.411 | 139.478 | 371.381 | 11.968 | 6.001 | 11.836 | 13.347 | ... | NaN | NaN | 2.566 | 0.079 | 2.882 | 3.742 | 1.539 | 1.891 | 0.931 | NaN |
| 36 | MPI-ESM-1-2-HAM | r1i1p1 | MPI-ESM-1-2-HAM_r1i1p1 | 1.935 | 139.333 | 182.547 | 13.682 | 9.330 | 11.438 | 11.509 | ... | 5.209 | 2.242 | 2.071 | 0.034 | 2.339 | 3.703 | 1.602 | 1.835 | 0.942 | 35.808 |
| 37 | MPI-ESM1-2-HR | r1i1p1 | MPI-ESM1-2-HR_r1i1p1 | 1.558 | 139.185 | 175.940 | 11.483 | 7.972 | 8.214 | 9.257 | ... | 1.855 | 1.226 | 1.518 | 0.029 | 1.667 | 3.610 | 1.464 | 1.869 | 0.785 | NaN |
| 38 | MPI-ESM1-2-LR | r1i1p1 | MPI-ESM1-2-LR_r1i1p1 | 1.586 | 139.240 | 169.975 | 12.861 | 7.665 | 9.964 | 9.711 | ... | 3.753 | 1.650 | 1.675 | 0.031 | 1.981 | 3.258 | 1.405 | 1.842 | 0.879 | 25.125 |
| 39 | MRI-ESM2-0 | r1i1p1 | MRI-ESM2-0_r1i1p1 | 1.542 | 139.105 | 262.614 | 12.888 | 7.231 | 9.446 | 9.306 | ... | NaN | NaN | 1.619 | 0.033 | 1.820 | 5.641 | 2.041 | 2.302 | 0.942 | NaN |
| 40 | NESM3 | r1i1p1 | NESM3_r1i1p1 | 1.958 | 139.286 | 252.038 | 16.135 | 11.223 | 13.198 | 15.035 | ... | 3.635 | 2.314 | 2.673 | 0.040 | 2.878 | 5.600 | 2.080 | 2.459 | 1.177 | 33.842 |
| 41 | NorCPM1 | r1i1p1 | NorCPM1_r1i1p1 | 1.562 | 139.040 | 302.481 | 16.905 | 10.218 | 13.985 | 13.010 | ... | 4.101 | 2.341 | 2.775 | 0.039 | 3.017 | 3.423 | 2.401 | 2.197 | 1.290 | 27.684 |
| 42 | NorESM2-MM | r1i1p1 | NorESM2-MM_r1i1p1 | 0.995 | 139.146 | 187.760 | 11.090 | 6.325 | 8.893 | 7.007 | ... | NaN | NaN | 1.900 | 0.032 | 2.038 | 3.128 | 1.273 | NaN | 0.828 | 19.449 |
| 43 | SAM0-UNICON | r1i1p1 | SAM0-UNICON_r1i1p1 | 1.444 | 139.142 | 238.562 | 13.340 | 9.123 | 11.336 | 10.244 | ... | 3.560 | 1.479 | 2.478 | 0.036 | 2.609 | 3.293 | 1.736 | 2.036 | 0.939 | 23.985 |
| 44 | TaiESM1 | r1i1p1 | TaiESM1_r1i1p1 | 1.335 | 139.102 | 211.710 | 11.730 | 8.043 | 10.044 | 8.601 | ... | NaN | NaN | 2.243 | 0.044 | 2.406 | NaN | 1.508 | NaN | 0.814 | NaN |
| 45 | my_model | r1i1p1 | my_model_r1i1p1 | 1.359 | NaN | 266.107 | 13.913 | 6.382 | 10.892 | 7.969 | ... | 2.906 | 26.545 | 2.090 | NaN | 2.276 | 3.033 | 1.508 | 1.577 | 0.793 | 24.472 |
46 rows × 28 columns
Customize variables to show (optional)
[15]:
# customize variables shown in portrait plot
#var_list = ["pr", "psl", "rltcre", "rlut", "rstcre", "rsut", "ta-200", "ta-850", "tas", "ts",
# "ua-200", "ua-850", "va-200", "va-850", "zg-500"]
var_list = sorted(var_list)
var_list.remove('sfcWind') # temporarliy removed sfc wind due to problem in observation
[16]:
# Simple re-order variables
if 'zg-500' in var_list and 'sfcWind' in var_list:
var_list.remove('zg-500')
idx_sfcWind = var_list.index('sfcWind')
var_list.insert(idx_sfcWind+1, 'zg-500')
print("var_list:", var_list)
var_list: ['pr', 'prw', 'psl', 'rlds', 'rltcre', 'rlus', 'rlut', 'rlutcs', 'rsds', 'rsdscs', 'rsdt', 'rstcre', 'rsut', 'rsutcs', 'ta-200', 'ta-850', 'tas', 'tauu', 'ts', 'ua-200', 'ua-850', 'va-200', 'va-850', 'zg-500']
Prepare input for portrait plot plotting function
[17]:
data_djf = df_dict['rms_xy']['djf']['global'][var_list].to_numpy()
data_mam = df_dict['rms_xy']['mam']['global'][var_list].to_numpy()
data_jja = df_dict['rms_xy']['jja']['global'][var_list].to_numpy()
data_son = df_dict['rms_xy']['son']['global'][var_list].to_numpy()
model_names = df_dict['rms_xyt']['ann']['global']['model'].tolist()
data_all = np.stack([data_djf, data_mam, data_jja, data_son])
print('data.shape:', data_all.shape)
print('len(var_list): ', len(var_list))
print('len(model_names): ', len(model_names))
xaxis_labels = var_list
yaxis_labels = model_names
data.shape: (4, 46, 24)
len(var_list): 24
len(model_names): 46
1.5 Normalize each column by its median for portrait plot
Use normalize_by_median function.
Parameters
data: 2d numpy arrayaxis: 0 (normalize each column) or 1 (normalize each row), default=0
Return
data_nor: 2d numpy array
[18]:
from pcmdi_metrics.graphics import normalize_by_median
data_djf_nor = normalize_by_median(data_djf)
data_mam_nor = normalize_by_median(data_mam)
data_jja_nor = normalize_by_median(data_jja)
data_son_nor = normalize_by_median(data_son)
[19]:
data_all_nor = np.stack([data_djf_nor, data_mam_nor, data_jja_nor, data_son_nor])
data_all_nor.shape
[19]:
(4, 46, 24)
2. Portrait Plot
Use Matplotlib-based PMP Visualization Function. Detailed description for the functions parameters and returns can be found in the API documentation.
[20]:
from pcmdi_metrics.graphics import portrait_plot
Portrait Plot with 4 Triangles (4 seasons)
data order is clockwise from top: top, right, bottom, left
[21]:
fig, ax, cbar = portrait_plot(data_all_nor,
xaxis_labels=xaxis_labels,
yaxis_labels=yaxis_labels,
cbar_label='RMSE',
box_as_square=True,
vrange=(-0.5, 0.5),
figsize=(15, 18),
cmap='RdYlBu_r',
cmap_bounds=[-0.5, -0.4, -0.3, -0.2, -0.1, 0, 0.1, 0.2, 0.3, 0.4, 0.5],
cbar_kw={"extend": "both"},
missing_color='grey',
legend_on=True,
legend_labels=['DJF', 'MAM', 'JJA', 'SON'],
legend_box_xy=(1.25, 1),
legend_box_size=3,
legend_lw=1,
legend_fontsize=12.5,
logo_rect = [0.85, 0.15, 0.07, 0.07]
)
ax.set_xticklabels(xaxis_labels, rotation=45, va='bottom', ha="left")
# Add title
ax.set_title("Seasonal climatology RMSE", fontsize=30, pad=30)
# Add data info
fig.text(1.25, 0.9, 'Data version\n'+data_version, transform=ax.transAxes,
fontsize=12, color='black', alpha=0.6, ha='left', va='top',)
# Add Watermark
ax.text(0.5, 0.5, 'Example', transform=ax.transAxes,
fontsize=100, color='black', alpha=0.5,
ha='center', va='center', rotation=25)
[21]:
Text(0.5, 0.5, 'Example')
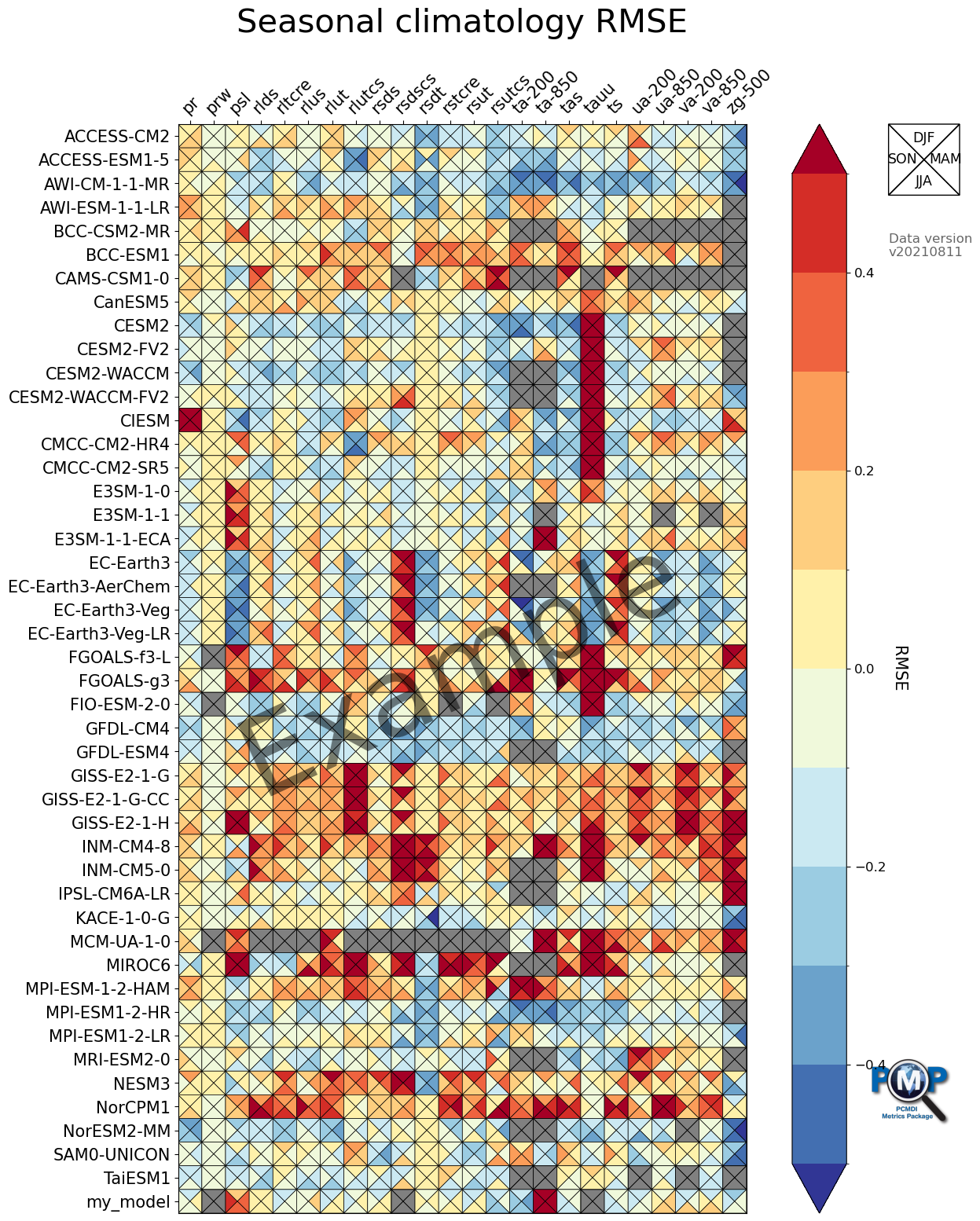
[22]:
# Save figure as an image file
fig.savefig('mean_clim_portrait_plot_4seasons_example_user_model.png', facecolor='w', bbox_inches='tight')
3. Parallel Coordinate Plot
[23]:
data = df_dict['rms_xyt']['ann']['global'][var_list].to_numpy()
model_names = df_dict['rms_xyt']['ann']['global']['model'].tolist()
#metric_names = ['\n['.join(var_unit.split(' [')) for var_unit in var_unit_list]
metric_names = var_list
model_highlights = ['my_model']
print('data.shape:', data.shape)
print('len(metric_names): ', len(metric_names))
print('len(model_names): ', len(model_names))
data.shape: (46, 24)
len(metric_names): 24
len(model_names): 46
[24]:
units_all = 'prw [kg m-2], pr [mm d-1], psl [Pa], rlds [W m-2], rsdscs [W m-2], rltcre [W m-2], rlus [W m-2], rlut [W m-2], rlutcs [W m-2], rsds [W m-2], rsdt [W m-2], rstcre [W m-2], rsus [W m-2], rsut [W m-2], rsutcs [W m-2], sfcWind [m s-1], zg-500 [m], ta-200 [K], ta-850 [K], tas [K], ts [K], ua-200 [m s-1], ua-850 [m s-1], uas [m s-1], va-200 [m s-1], va-850 [m s-1], vas [m s-1], tauu [Pa]'
units_all.split(', ')
var_unit_list = []
for var in var_list:
found = False
for var_units in units_all.split(', '):
tmp1 = var_units.split(' [')[0]
#print(var, tmp1)
if tmp1 == var:
unit = '[' + var_units.split(' [')[1]
var_unit_list.append(var + '\n' + unit)
found = True
break
if found is False:
print(var, 'not found')
print('var_unit_list:', var_unit_list)
metric_names = var_unit_list
var_unit_list: ['pr\n[mm d-1]', 'prw\n[kg m-2]', 'psl\n[Pa]', 'rlds\n[W m-2]', 'rltcre\n[W m-2]', 'rlus\n[W m-2]', 'rlut\n[W m-2]', 'rlutcs\n[W m-2]', 'rsds\n[W m-2]', 'rsdscs\n[W m-2]', 'rsdt\n[W m-2]', 'rstcre\n[W m-2]', 'rsut\n[W m-2]', 'rsutcs\n[W m-2]', 'ta-200\n[K]', 'ta-850\n[K]', 'tas\n[K]', 'tauu\n[Pa]', 'ts\n[K]', 'ua-200\n[m s-1]', 'ua-850\n[m s-1]', 'va-200\n[m s-1]', 'va-850\n[m s-1]', 'zg-500\n[m]']
[25]:
df_dict['rms_xyt']['ann']['global'][var_list].columns
[25]:
Index(['pr', 'prw', 'psl', 'rlds', 'rltcre', 'rlus', 'rlut', 'rlutcs', 'rsds',
'rsdscs', 'rsdt', 'rstcre', 'rsut', 'rsutcs', 'ta-200', 'ta-850', 'tas',
'tauu', 'ts', 'ua-200', 'ua-850', 'va-200', 'va-850', 'zg-500'],
dtype='object')
Use parallel coordinate plot function of PMP
Detailed description for the functions parameters and returns can be found in the API documentation.
[26]:
from pcmdi_metrics.graphics import parallel_coordinate_plot
[27]:
fig, ax = parallel_coordinate_plot(data, metric_names, model_names, model_highlights,
title='Mean Climate: RMS_XYT, ANN, Global',
figsize=(21, 7),
colormap='tab20',
show_boxplot=False,
show_violin=True,
xtick_labelsize=10,
logo_rect=[0.8, 0.8, 0.15, 0.15])
#fig.text(0.99, -0.45, 'Data version\n'+data_version, transform=ax.transAxes,
# fontsize=12, color='black', alpha=0.6, ha='right', va='bottom',)
# Add Watermark
ax.text(0.5, 0.5, 'Example', transform=ax.transAxes,
fontsize=100, color='black', alpha=0.2,
ha='center', va='center', rotation=25)
[27]:
Text(0.5, 0.5, 'Example')
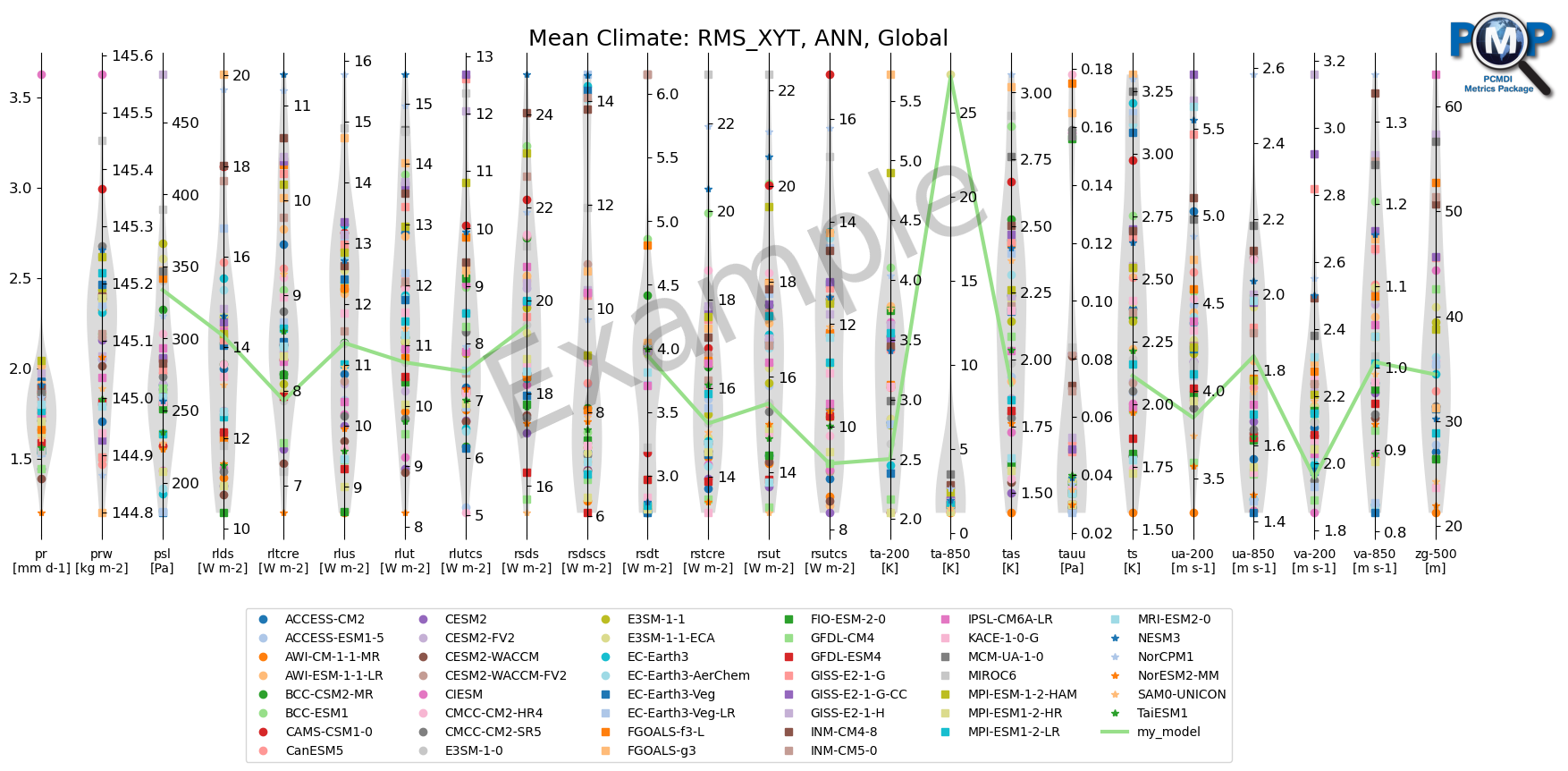
[28]:
# Save figure as an image file
fig.savefig('mean_clim_parallel_coordinate_plot_example_user_model.png', facecolor='w', bbox_inches='tight')
4. Box plots
Generate a set of box plots using `matplotlib <https://matplotlib.org/>`__ and `pandas.DataFrame.boxplot <https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.boxplot.html>`__.
Note: The box extends from the first quartile (Q1) to the third quartile (Q3) of the data, with a line at the median. The whiskers extend from the box by 1.5x the inter-quartile range (IQR). Flier points are those past the end of the whiskers. See https://matplotlib.org/3.5.0/api/_as_gen/matplotlib.pyplot.boxplot.html and https://en.wikipedia.org/wiki/Box_plot for reference.
[29]:
stat = 'rms_xyt'
season = 'ann'
region = 'global'
[30]:
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import math
fig = plt.figure(figsize=(14,14))
ncols = 5
nrows = int(math.ceil(len(var_list)/ncols))
for index, var in enumerate(var_list):
fig.add_subplot(nrows, ncols, index+1)
# Box plot for library (i.e., CMIP6 models)
ax = library.df_dict[stat][season][region].boxplot(
[var],
# Customize box plot
grid=False, fontsize=16,
color=dict(boxes='black', whiskers='black', medians='black', caps='black'),
boxprops=dict(linestyle='-', linewidth=1.5),
flierprops=dict(linestyle='-', linewidth=1.5),
medianprops=dict(linestyle='-', linewidth=1.5, color='black'),
whiskerprops=dict(linestyle='-', linewidth=1.5),
capprops=dict(linestyle='-', linewidth=1.5),
showfliers=False, # mute showing outliers
rot=0,
)
# Add marker for test case (i.e. user models)
try:
my_model = test_case.df_dict[stat][season][region][var]
ax.plot(1, my_model, 'o', c='red', markersize=10, label='my_model')
except:
pass
# Show unit as y-axis label
ax.set_ylabel(var_unit_list[index].split('[')[-1].split(']')[0])
# Show legend at upper right corner of the figure
if index == ncols-1:
h, l = ax.get_legend_handles_labels()
black_patch = mpatches.Patch(color='black', label='CMIP6', fill=False)
ax.legend(handles= [black_patch] + h,
bbox_to_anchor=(1, 1.02), loc='lower right', ncol=2)
# Add Watermark
fig.text(0.5, 0.4, 'Example',
fontsize=100, color='black', alpha=0.1,
ha='center', va='center', rotation=25)
fig.suptitle('rms_xyt, ann, global', fontsize=20)
fig.tight_layout(pad=1.5)
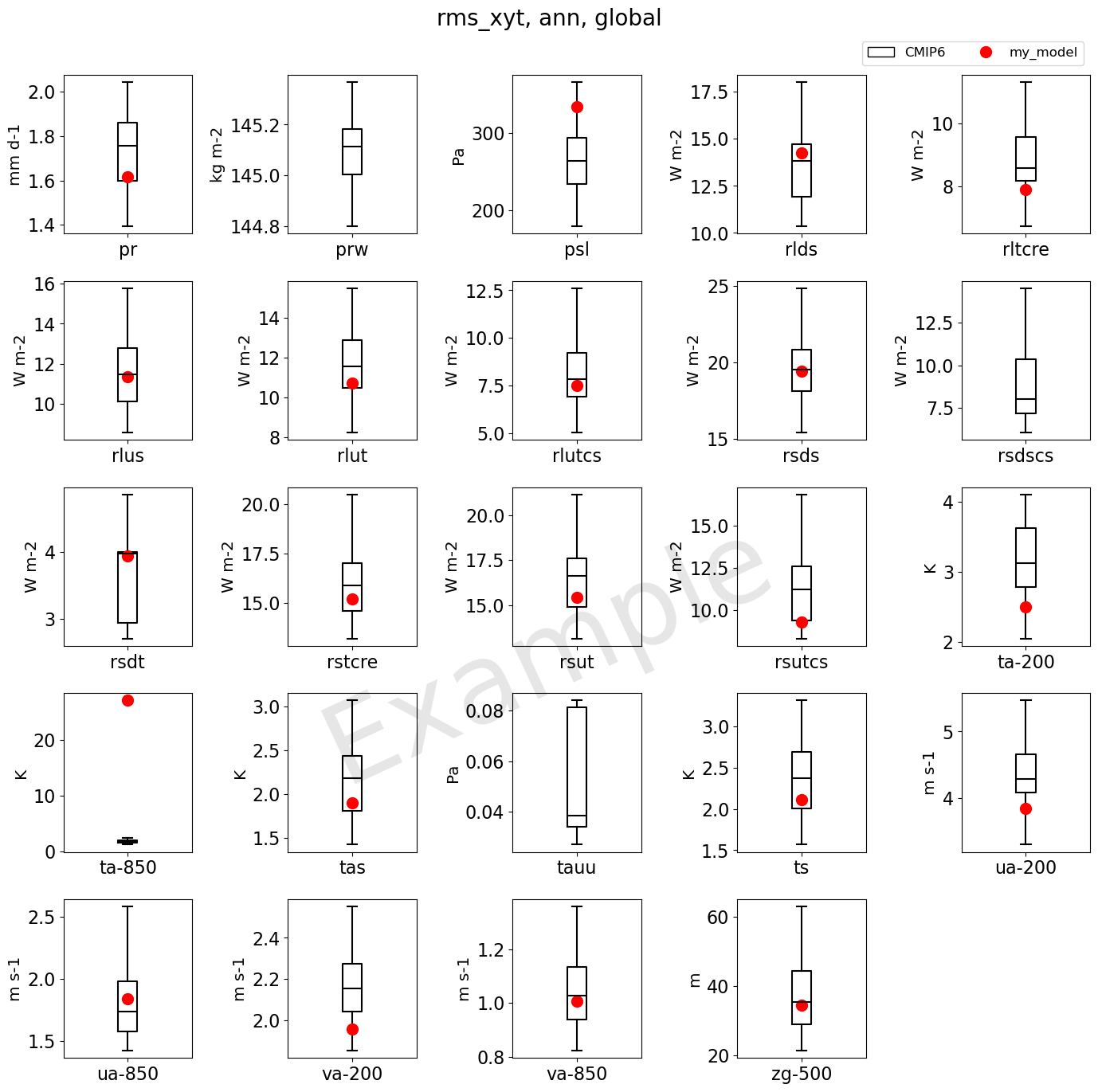
[31]:
fig.savefig('mean_clim_box_plot_example_user_model.png', facecolor='w', bbox_inches='tight')
5. Taylor Diagram
Detailed description for the functions parameters and returns can be found in the API documentation.
[32]:
from pcmdi_metrics.graphics import TaylorDiagram
Select variabile, season, and region for Taylor Diagram
[33]:
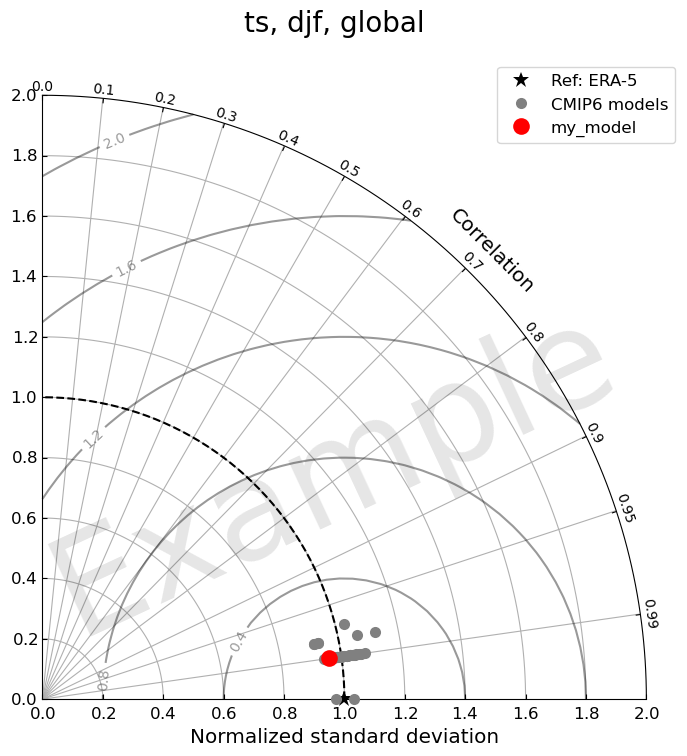
var = "ts"
season = "djf"
region = "global"
5.1 Identify all models
[34]:
fig = plt.figure(figsize=(8,8))
stddev = combined.df_dict["std_xy"][season][region][var].to_numpy()
refstd = combined.df_dict['std-obs_xy'][season][region][var][0]
corrcoeff = combined.df_dict["cor_xy"][season][region][var].to_numpy()
models = combined.df_dict["cor_xy"][season][region]['model'].to_list()
colors = plt.matplotlib.cm.jet(np.linspace(0, 1, len(models)))
fig, ax = TaylorDiagram(stddev, corrcoeff, refstd, fig, colors=colors, normalize=True, labels=models, ref_label='Ref: '+var_ref_dict[var])
ax.legend(bbox_to_anchor=(1.05, 0), loc='lower left', ncol=2)
fig.suptitle(', '.join([var, season, region]), fontsize=20)
# Add Watermark
fig.text(0.5, 0.4, 'Example',
fontsize=100, color='black', alpha=0.1,
ha='center', va='center', rotation=25)
[34]:
Text(0.5, 0.4, 'Example')
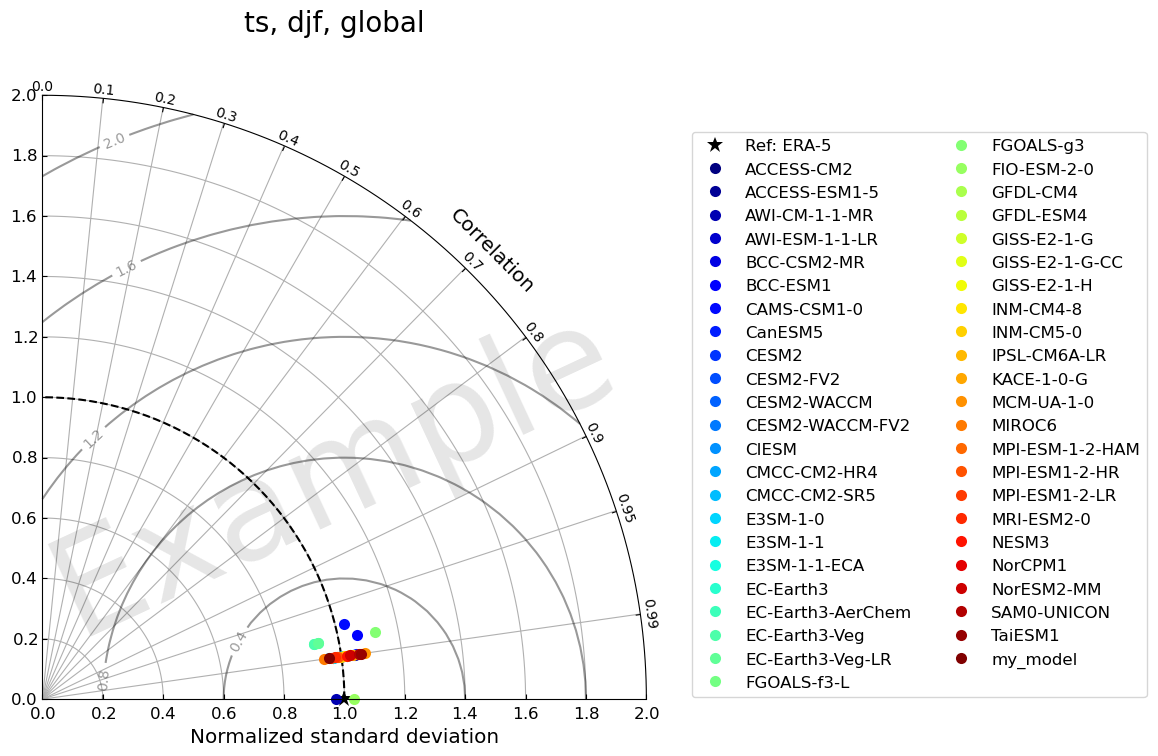
5.2 Highlight user’s model
[35]:
fig = plt.figure(figsize=(8,8))
# Customize plot to highlight user's model in red, others in grey
colors = list()
labels = list()
markers = list()
markersizes = list()
zorders = list()
for i, model in enumerate(models):
if i == len(models)-1:
labels.append(model)
colors.append('red')
markers.append('o')
markersizes.append(12)
zorders.append(100)
else:
if i == 0:
labels.append('CMIP6 models')
else:
labels.append(None)
colors.append('grey')
markersizes.append(8)
markers.append('o')
zorders.append(10)
fig, ax = TaylorDiagram(stddev, corrcoeff, refstd, fig, colors=colors, normalize=True,
labels=labels, markers=markers,
markersizes=markersizes, zorders=zorders, ref_label='Ref: '+var_ref_dict[var])
ax.legend(bbox_to_anchor=(1.05, 1.05), loc='upper right', ncol=1)
fig.suptitle(', '.join([var, season, region]), fontsize=20)
# Add Watermark
fig.text(0.5, 0.4, 'Example',
fontsize=100, color='black', alpha=0.1,
ha='center', va='center', rotation=25)
[35]:
Text(0.5, 0.4, 'Example')
[36]:
fig.savefig('mean_clim_taylor_diagram_example_user_model.png', facecolor='w', bbox_inches='tight')