Taylor Diagram: Mean Climate, comparing CMIP5 & CMIP6 models
Visualize PMP’s Mean climate metrics output using PMP’s Taylor Diagram. This notebook generates a static image of Taylor Diagram for mean climate metrics obtained from CMIP5 and CMIP6 models, and compare multi-model averaged statistics from each group.
Written by Jiwoo Lee (LLNL/PCMDI)
Last update: 2022. 10
1. Read data from JSON files
Input data for portrait plot is expected as a set a (stacked or list of) 2-d numpy array(s) with list of strings for x and y axes labels.
1.1 Download PMP output JSON files for CMIP models
[1]:
import glob
import os
import numpy as np
import requests
import pandas as pd
from pcmdi_metrics.graphics import download_archived_results
PMP output files downloadable from the PMP results archive.
[2]:
vars = ['pr', 'prw', 'psl', 'rlds', 'rltcre', 'rlus', 'rlut', 'rlutcs', 'rsds', 'rsdscs', 'rsdt', 'rstcre', 'rsut', 'rsutcs', 'sfcWind',
'ta-200', 'ta-850', 'tas', 'tauu', 'ts', 'ua-200', 'ua-850', 'va-200', 'va-850', 'zg-500']
[3]:
json_dir = './json_files'
[4]:
mip = "cmip5"
exp = "historical"
data_version = "v20220928"
[5]:
for var in vars:
path = "metrics_results/mean_climate/"+mip+"/"+exp+"/"+data_version+"/"+var+"."+mip+"."+exp+".regrid2.2p5x2p5."+data_version+".json"
if not os.path.exists(os.path.join(json_dir, path.split('/')[-1])):
download_archived_results(path, json_dir)
[6]:
mip = "cmip6"
exp = "historical"
data_version = "v20220928"
[7]:
for var in vars:
path = "metrics_results/mean_climate/"+mip+"/"+exp+"/"+data_version+"/"+var+"."+mip+"."+exp+".regrid2.2p5x2p5."+data_version+".json"
if not os.path.exists(os.path.join(json_dir, path.split('/')[-1])):
download_archived_results(path, json_dir)
Check JSON files
[8]:
json_list_1 = sorted(glob.glob(os.path.join(json_dir, '*.cmip5.' + exp + '*' + data_version + '.json')))
json_list_2 = sorted(glob.glob(os.path.join(json_dir, '*.cmip6.' + exp + '*' + data_version + '.json')))
[9]:
print('CMIP5 JSON files:')
for i, json_file in enumerate(json_list_1):
print(i+1, json_file.split('/')[-1])
print('CMIP6 JSON files:')
for i, json_file in enumerate(json_list_2):
print(i+1, json_file.split('/')[-1])
CMIP5 JSON files:
1 pr.cmip5.historical.regrid2.2p5x2p5.v20220928.json
2 prw.cmip5.historical.regrid2.2p5x2p5.v20220928.json
3 psl.cmip5.historical.regrid2.2p5x2p5.v20220928.json
4 rlds.cmip5.historical.regrid2.2p5x2p5.v20220928.json
5 rltcre.cmip5.historical.regrid2.2p5x2p5.v20220928.json
6 rlus.cmip5.historical.regrid2.2p5x2p5.v20220928.json
7 rlut.cmip5.historical.regrid2.2p5x2p5.v20220928.json
8 rlutcs.cmip5.historical.regrid2.2p5x2p5.v20220928.json
9 rsds.cmip5.historical.regrid2.2p5x2p5.v20220928.json
10 rsdscs.cmip5.historical.regrid2.2p5x2p5.v20220928.json
11 rsdt.cmip5.historical.regrid2.2p5x2p5.v20220928.json
12 rstcre.cmip5.historical.regrid2.2p5x2p5.v20220928.json
13 rsut.cmip5.historical.regrid2.2p5x2p5.v20220928.json
14 rsutcs.cmip5.historical.regrid2.2p5x2p5.v20220928.json
15 sfcWind.cmip5.historical.regrid2.2p5x2p5.v20220928.json
16 ta-200.cmip5.historical.regrid2.2p5x2p5.v20220928.json
17 ta-850.cmip5.historical.regrid2.2p5x2p5.v20220928.json
18 tas.cmip5.historical.regrid2.2p5x2p5.v20220928.json
19 tauu.cmip5.historical.regrid2.2p5x2p5.v20220928.json
20 ts.cmip5.historical.regrid2.2p5x2p5.v20220928.json
21 ua-200.cmip5.historical.regrid2.2p5x2p5.v20220928.json
22 ua-850.cmip5.historical.regrid2.2p5x2p5.v20220928.json
23 va-200.cmip5.historical.regrid2.2p5x2p5.v20220928.json
24 va-850.cmip5.historical.regrid2.2p5x2p5.v20220928.json
25 zg-500.cmip5.historical.regrid2.2p5x2p5.v20220928.json
CMIP6 JSON files:
1 pr.cmip6.historical.regrid2.2p5x2p5.v20220928.json
2 prw.cmip6.historical.regrid2.2p5x2p5.v20220928.json
3 psl.cmip6.historical.regrid2.2p5x2p5.v20220928.json
4 rlds.cmip6.historical.regrid2.2p5x2p5.v20220928.json
5 rltcre.cmip6.historical.regrid2.2p5x2p5.v20220928.json
6 rlus.cmip6.historical.regrid2.2p5x2p5.v20220928.json
7 rlut.cmip6.historical.regrid2.2p5x2p5.v20220928.json
8 rlutcs.cmip6.historical.regrid2.2p5x2p5.v20220928.json
9 rsds.cmip6.historical.regrid2.2p5x2p5.v20220928.json
10 rsdscs.cmip6.historical.regrid2.2p5x2p5.v20220928.json
11 rsdt.cmip6.historical.regrid2.2p5x2p5.v20220928.json
12 rstcre.cmip6.historical.regrid2.2p5x2p5.v20220928.json
13 rsut.cmip6.historical.regrid2.2p5x2p5.v20220928.json
14 rsutcs.cmip6.historical.regrid2.2p5x2p5.v20220928.json
15 sfcWind.cmip6.historical.regrid2.2p5x2p5.v20220928.json
16 ta-200.cmip6.historical.regrid2.2p5x2p5.v20220928.json
17 ta-850.cmip6.historical.regrid2.2p5x2p5.v20220928.json
18 tas.cmip6.historical.regrid2.2p5x2p5.v20220928.json
19 tauu.cmip6.historical.regrid2.2p5x2p5.v20220928.json
20 ts.cmip6.historical.regrid2.2p5x2p5.v20220928.json
21 ua-200.cmip6.historical.regrid2.2p5x2p5.v20220928.json
22 ua-850.cmip6.historical.regrid2.2p5x2p5.v20220928.json
23 va-200.cmip6.historical.regrid2.2p5x2p5.v20220928.json
24 va-850.cmip6.historical.regrid2.2p5x2p5.v20220928.json
25 zg-500.cmip6.historical.regrid2.2p5x2p5.v20220928.json
1.2 Extract data from JSON files
Use Metrics class (that use read_mean_clim_json_files function underneath) to extract data from the above JSON files.
Parameters
json_list: list of string, where each element is for path/file for PMP output JSON files
Returned object includes
df_dict: dictionary that has[stat][season][region]hierarchy structure storing pandas dataframe for metric numbers (Rows: models, Columns: variables (i.e., 2d array)var_list: list of string, all variables from JSON filesvar_unit_list: list of string, all variables and its units from JSON filesvar_ref_dict: dictonary for reference dataset used for each variableregions: list of string, regionsstats: list of string, statistics
[10]:
from pcmdi_metrics.graphics import Metrics
[11]:
library_cmip5 = Metrics(json_list_1, mip="cmip5")
[12]:
library_cmip6 = Metrics(json_list_2, mip="cmip6")
1.3 Merge data
[13]:
# merge dataframes
combined = library_cmip5.merge(library_cmip6)
[14]:
var = "pr"
season = "jja"
region = "global"
1.4 Multi-model averaged statistics
1.4.1 Standard deviation
[15]:
stat1 = "std_xy"
# mean value of statistics from multi models in each CMIP
combined.df_dict[stat1][season][region].loc['CMIP5 mean'] = library_cmip5.df_dict[stat1][season][region].mean(numeric_only=True, skipna=True)
combined.df_dict[stat1][season][region].loc['CMIP6 mean'] = library_cmip6.df_dict[stat1][season][region].mean(numeric_only=True, skipna=True)
combined.df_dict[stat1][season][region].at['CMIP5 mean', 'model'] = 'CMIP5 mean'
combined.df_dict[stat1][season][region].at['CMIP6 mean', 'model'] = 'CMIP6 mean'
1.4.2 Correlation
Correlation cannot be simply averaged. The average code below follows this, which follows this.
[16]:
def average_cor(corr):
mean_z = np.nanmean(np.arctanh(corr).values)
mean_corr = np.tanh(mean_z)
return mean_corr
[17]:
stat2 = "cor_xy"
corr_cmip5 = library_cmip5.df_dict[stat2][season][region][var]
corr_cmip6 = library_cmip6.df_dict[stat2][season][region][var]
# mean value of correlation from multi models in each CMIP
corr_ave_cmip5 = average_cor(corr_cmip5)
corr_ave_cmip6 = average_cor(corr_cmip6)
[18]:
combined.df_dict[stat2][season][region].loc['CMIP5 mean', var] = corr_ave_cmip5
combined.df_dict[stat2][season][region].loc['CMIP6 mean', var] = corr_ave_cmip6
[19]:
combined.df_dict[stat2][season][region].at['CMIP5 mean', 'model'] = 'CMIP5 mean'
combined.df_dict[stat2][season][region].at['CMIP6 mean', 'model'] = 'CMIP6 mean'
1.5 Reference dataset
[20]:
var_ref_dict = combined.var_ref_dict
[21]:
var_ref_dict
[21]:
{'pr': 'GPCP-2-3',
'prw': 'REMSS-PRW-v07r01',
'psl': 'ERA-5',
'rlds': 'CERES-EBAF-4-1',
'rltcre': 'CERES-EBAF-4-1',
'rlus': 'CERES-EBAF-4-1',
'rlut': 'CERES-EBAF-4-1',
'rlutcs': 'CERES-EBAF-4-1',
'rsds': 'CERES-EBAF-4-1',
'rsdscs': 'CERES-EBAF-4-1',
'rsdt': 'CERES-EBAF-4-1',
'rstcre': 'CERES-EBAF-4-1',
'rsut': 'CERES-EBAF-4-1',
'rsutcs': 'CERES-EBAF-4-1',
'sfcWind': 'REMSS-PRW-v07r01',
'ta-200': 'ERA-5',
'ta-850': 'ERA-5',
'tas': 'ERA-5',
'tauu': 'ERA-INT',
'ts': 'ERA-5',
'ua-200': 'ERA-5',
'ua-850': 'ERA-5',
'va-200': 'ERA-5',
'va-850': 'ERA-5',
'zg-500': 'ERA-5'}
2. Plot
Usage of TaylorDiagram function can be found at taylor_diagram_example.ipynb
[22]:
import matplotlib.pyplot as plt
from pcmdi_metrics.graphics import TaylorDiagram
[23]:
stddev = combined.df_dict["std_xy"][season][region][var].to_numpy()
refstd = combined.df_dict['std-obs_xy'][season][region][var][0]
corrcoeff = combined.df_dict["cor_xy"][season][region][var].to_numpy()
models = combined.df_dict["cor_xy"][season][region]['model'].to_list()
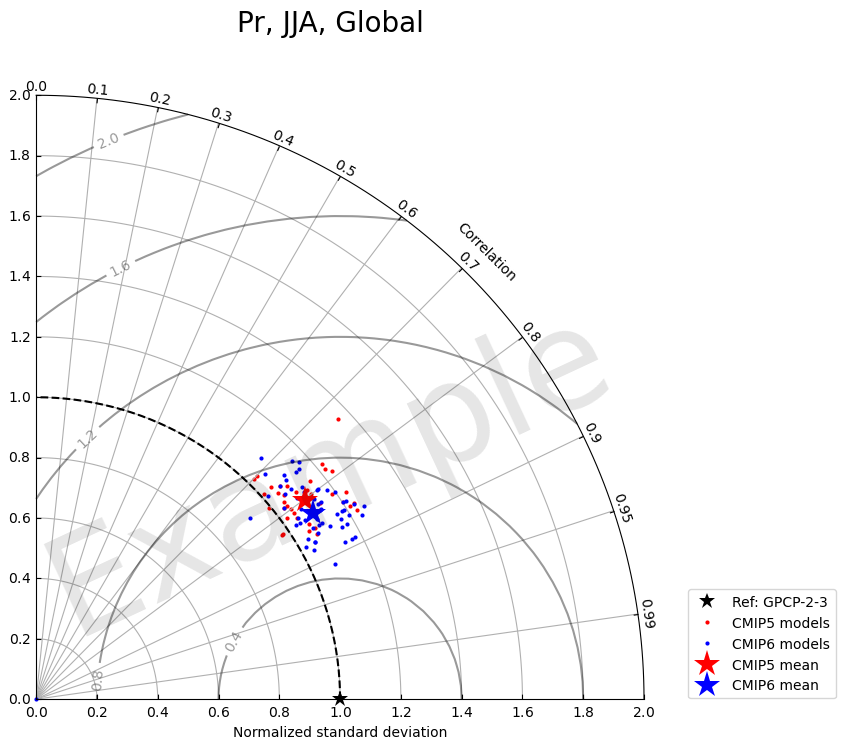
2.1 Plot example 1: label all models
[24]:
fig = plt.figure(figsize=(8,8))
# Customize plot
models1 = library_cmip5.df_dict["cor_xy"][season][region]['model'].to_list()
models2 = library_cmip6.df_dict["cor_xy"][season][region]['model'].to_list()
colors1 = plt.matplotlib.cm.YlOrBr_r(np.linspace(0.1, 0.9, len(models1))) # For CMIP5 models
colors2 = plt.matplotlib.cm.GnBu(np.linspace(0.1, 0.9, len(models2))) # For CMIP6 models
colors3 = plt.matplotlib.cm.bwr_r(np.linspace(0, 1, 2)) # For CMIP5 mean and CMIP6 mean
markers1 = ['o'] * len(models1) # For CMIP5 models
markers2 = ['s'] * len(models2) # For CMIP6 models
markers3 = ['*', '*'] # For CMIP5 mean and CMIP6 mean
markersizes1 = [8] * len(models1) # For CMIP5 models
markersizes2 = [8] * len(models2) # For CMIP6 models
markersizes3 = [15] * 2 # For CMIP5 mean and CMIP6 mean
zorders1 = [1] * len(models1) # For CMIP5 models
zorders2 = [2] * len(models2) # For CMIP6 models
zorders3 = [100, 101] # For CMIP5 mean and CMIP6 mean
# combine them and build a new colormap
colors = np.vstack((colors1, colors2, colors3))
markers = markers1 + markers2 + markers3
zorders = zorders1 + zorders2 + zorders3
markersizes = markersizes1 + markersizes2 + markersizes3
fig, ax = TaylorDiagram(stddev, corrcoeff, refstd,
fig=fig,
colors=colors,
normalize=True,
labels=models, ref_label='Ref: '+var_ref_dict[var],
markers=markers, zorders=zorders, markersizes=markersizes
)
ax.legend(bbox_to_anchor=(1.05, 0), loc='lower left', ncol=2)
fig.suptitle(', '.join([var.title(), season.upper(), region.title()]), fontsize=20)
# Add Watermark
fig.text(0.5, 0.4, 'Example',
fontsize=100, color='black', alpha=0.1,
ha='center', va='center', rotation='25')
[24]:
Text(0.5, 0.4, 'Example')
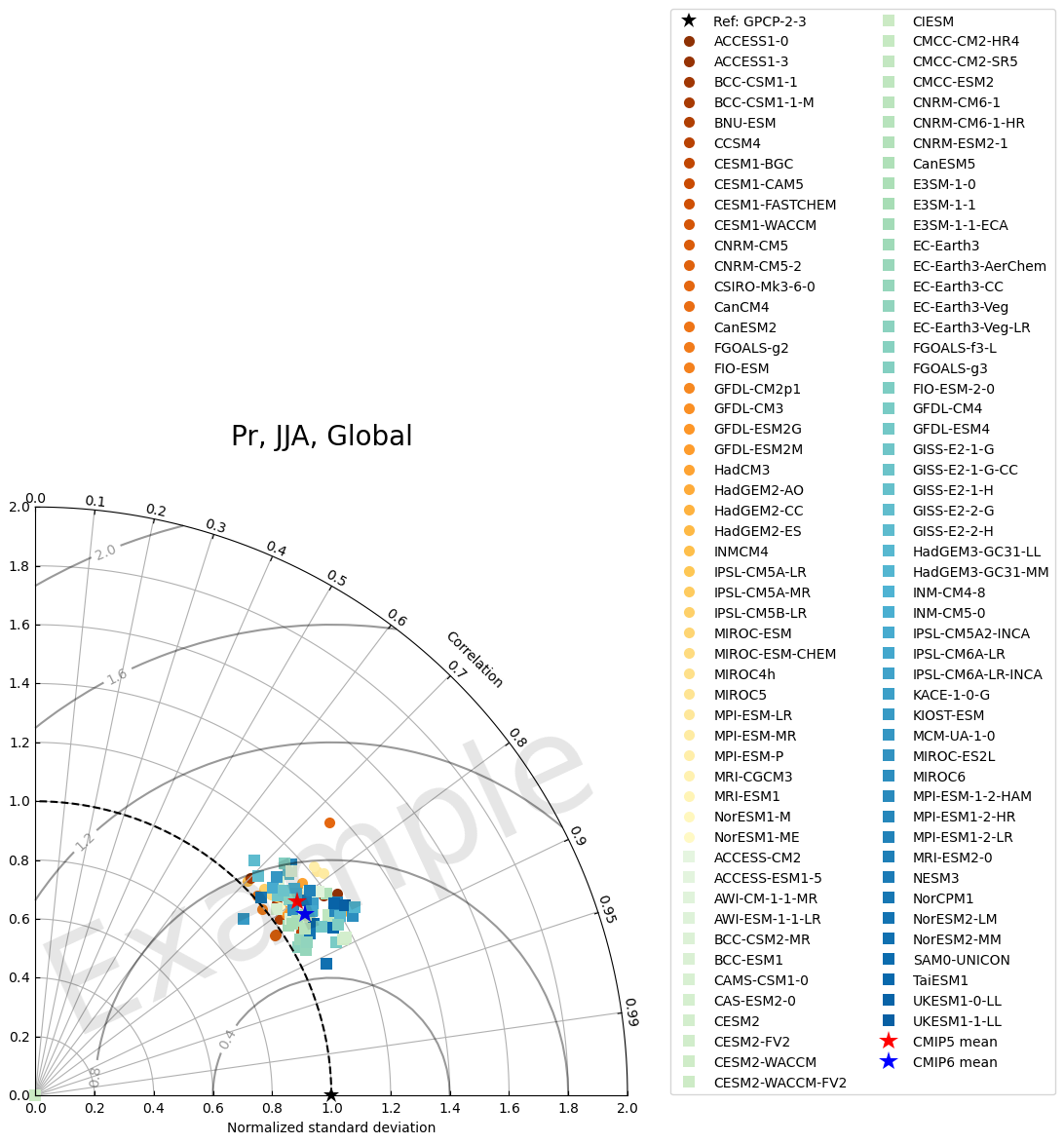
2.2 Plot example 2: label only model group
[25]:
fig = plt.figure(figsize=(8,8))
# Customize plot
models1 = library_cmip5.df_dict["cor_xy"][season][region]['model'].to_list()
models2 = library_cmip6.df_dict["cor_xy"][season][region]['model'].to_list()
colors1 = ['red'] * len(models1) # For CMIP5 models
colors2 = ['blue'] * len(models2) # For CMIP6 models
colors3 = ['red', 'blue'] # For CMIP5 mean and CMIP6 mean
markers1 = ['o'] * len(models1) # For CMIP5 models
markers2 = ['o'] * len(models2) # For CMIP6 models
markers3 = ['*', '*'] # For CMIP5 mean and CMIP6 mean
markersizes1 = [3] * len(models1) # For CMIP5 models
markersizes2 = [3] * len(models2) # For CMIP6 models
markersizes3 = [20] * 2 # For CMIP5 mean and CMIP6 mean
zorders1 = [1] * len(models1) # For CMIP5 models
zorders2 = [2] * len(models2) # For CMIP6 models
zorders3 = [100, 101] # For CMIP5 mean and CMIP6 mean
labels1 = ['CMIP5 models'] + [None] * (len(models1) - 1) # For CMIP5 models
labels2 = ['CMIP6 models'] + [None] * (len(models2) - 1) # For CMIP6 models
labels3 = ['CMIP5 mean', 'CMIP6 mean'] # For CMIP5 mean and CMIP6 mean
# combine them and build a new colormap
colors = colors1 + colors2 + colors3
markers = markers1 + markers2 + markers3
zorders = zorders1 + zorders2 + zorders3
labels = labels1 + labels2 + labels3
markersizes = markersizes1 + markersizes2 + markersizes3
fig, ax = TaylorDiagram(stddev, corrcoeff, refstd, fig=fig,
colors=colors, normalize=True,
labels=labels, ref_label='Ref: '+var_ref_dict[var],
markers=markers, zorders=zorders, markersizes=markersizes
)
ax.legend(bbox_to_anchor=(1.05, 0), loc='lower left', ncol=1)
fig.suptitle(', '.join([var.title(), season.upper(), region.title()]), fontsize=20)
# Add Watermark
fig.text(0.5, 0.4, 'Example',
fontsize=100, color='black', alpha=0.1,
ha='center', va='center', rotation='25')
[25]:
Text(0.5, 0.4, 'Example')
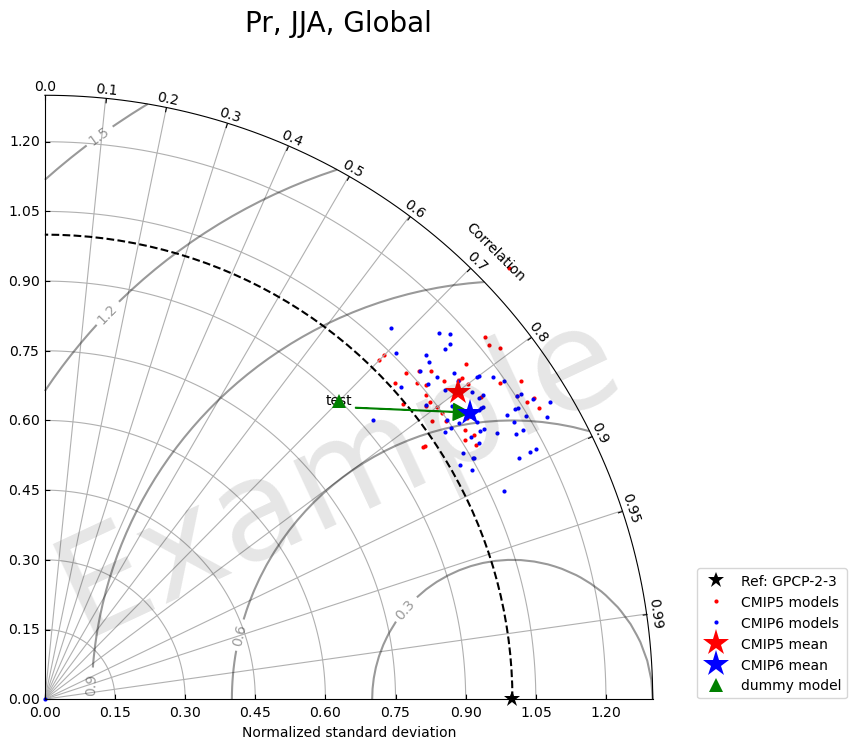
2.3 Plot example 3: Add model-comparing arrow
[26]:
fig = plt.figure(figsize=(8,8))
# Add dummy data
colors.append('green')
markers.append('^')
zorders.append(100)
labels.append('dummy model')
markersizes.append(10)
stddev_tmp = np.append(stddev, [refstd*0.9])
corrcoeff_tmp = np.append(corrcoeff, [0.7])
arrowprops_dict = dict(color='green',
lw=1,
width=0.5,
shrink=0,
zorder=100,
)
fig, ax = TaylorDiagram(stddev_tmp, corrcoeff_tmp, refstd,
fig=fig,
colors=colors, normalize=True,
labels=labels, ref_label='Ref: '+var_ref_dict[var],
markers=markers, zorders=zorders, markersizes=markersizes,
compare_models=[('dummy model', 'CMIP6 mean')],
arrowprops_dict=arrowprops_dict,
smax=1.3,
annotate_text='test',
)
ax.legend(bbox_to_anchor=(1.05, 0), loc='lower left', ncol=1)
fig.suptitle(', '.join([var.title(), season.upper(), region.title()]), fontsize=20)
# Add Watermark
fig.text(0.5, 0.4, 'Example',
fontsize=100, color='black', alpha=0.1,
ha='center', va='center', rotation='25')
[26]:
Text(0.5, 0.4, 'Example')